A locked nucleic acid (LNA), also known as bridged nucleic acid (BNA),[1] and often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation, which is often found in the A-form duplexes. This structure provides for increased stability against enzymatic degradation.[2][3][4][5] LNA also offers improved specificity and affinity in base-pairing as a monomer or a constituent of an oligonucleotide.[6] LNA nucleotides can be mixed with DNA or RNA residues in a oligonucleotide.
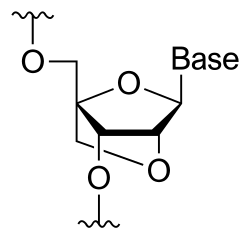
Synthesis
editObika et al. were the first to chemically synthesize LNA in 1997,[7] independently followed by Jesper Wengel's group in 1998.[8] This became possible after Zamecnick and Stephenson laid the groundwork on the possibility of oligonucleotides being great agents for controlling gene expression in 1978.[9] To date, two different approaches, referred to as linear and convergent strategies respectively, have been shown to produce high yield and efficient LNAs. The linear strategy of synthesis was first detailed in the works of Obika et al.[7] In this approach, uridine (or any readily available RNA nucleoside) can be used as the starting material. The convergent strategy requires the synthesis of a sugar intermediate which serves a glycosyl donor necessary for coupling with nucleobases. Commonly, D-glucose is used to produce the sugar intermediate which is subsequently reacted with nucleobases using a modified Vorbrügen procedure allowing for stereoselective coupling.[10]
The addition of different moieties has remained a possibility with the maintenance of key physicochemical properties like the high affinity and specificity evident in the originally synthesized LNA.[8] Such oligomers are synthesized chemically and are commercially available.
Incorporation into DNA/RNA
editLNA can be incorporated into DNA and RNA using the promiscuity of certain DNA and RNA polymerases. Phusion DNA polymerase, a commercially designed enzyme based on a Pfu DNA polymerase, efficiently incorporates LNA into DNA.[11]
Properties
editLNA offers enhanced biostability compared to biological nucleic acids. LNA modified oligonucleotides have demonstrated improved thermodynamics in hybridization to RNA, ssDNA, and dsDNA.[11]
Applications
editLNAzymes
editDNAzymes can be modified to include LNA residues, producing LNAzymes (LNA-modified DNAzymes). These modified oligonucleotides, like their DNAzyme relatives, are generally endonucleases that bind to specific RNA target sequences and cleave the phosphodiester bond that exists between the nucleotides.[12] However, they demonstrate more efficient cleavage of phosphodiester bonds compared to their unmodified counterparts. [13] Modification of the substrate recognition arms of DNAzymes with LNA monomers yields a LNAzyme which recognizes coxsackievirus A21 (CAV-21) and cleaves its RNA target sequence similar to one in the 5' untranslated region (5' UTR) of the human rhinovirus-14 (HRV-14); a sequence unrecognized by unmodified DNAzymes.[14]
Therapeutics
editUsing LNA based oligonucleotides therapeutically is an emerging field in biotechnology.[15] A variety of LNA oligonucleotides have been assessed for their pharmacokinetic and toxicity profiles. Studies concluded that LNA toxicity is generally independent of oligonucleotide sequence, and displays a preferential safety profile for translatable therapeutic applications.[8]
LNA has been investigated for its therapeutic properties in treating cancers and infectious diseases. A locked nucleic acid phosphorothioate antisense molecule, termed SPC2996, has been developed to target the mRNA coding for Bcl-2 oncoprotein, a protein that inhibits apoptosis in chronic lymphocytic leukemia cells (CLL). Phase I and II clinical trials demonstrated a dose dependent reduction in circulating CLL cells in approximately 30% of the sample population, suggesting further investigation into SPC2996.[16]
LNA has also been applied to Miravirsen, an experimental therapeutic intended for the treatment of Hepatitis C, constituting a 15-nucleotide phosphorothioate sequence with binding specificity for MiR-122 (a miRNA expressed in hepatocytes).[17][18]
Detection and diagnosis
editAllele-specific PCR using LNA allows for the design of shorter primers, without compromising binding specificity.[19]
LNA has been incorporated in fluorescence in situ hybridization (FISH).[20] FISH is a common technique used to visualize genetic material in a variety of cells, but studies noted that this technique has been limited by low probe hybridization efficiency. Conversely, LNA-incorporated probes demonstrated increased hybridization efficiency in both DNA and RNA. The improved efficiency of LNA-incorporated FISH has resulted in FISH analysis of the human chromosome, several types of non-human cells, and microarrays.[20]
LNA genotyping assays have been conducted as well, specifically to detect a mutation in apolipoprotein B.[20]
For its high affinity for mismatch discrimination, LNA has been studied for its applications in diagnostic tools. Immobilized LNA probes have been introduced in a multiplex SNP genotyping assay.[15]
Gene editing
editLNA-modified ssODNs (synthetic single-stranded DNA oligonucleotides) can be used like ordinary ssODNs for single-base gene editing. Using LNA at or close to the intended site of modification offers evasion of DNA mismatch repair due to the higher thermodynamic stability it has.[21]
References
edit- ^ Elayadi, Anissa N.; Braasch, Dwaine A.; Corey, David R. (August 2002). "Implications of High-Affinity Hybridization by Locked Nucleic Acid Oligomers for Inhibition of Human Telomerase †". Biochemistry. 41 (31): 9973–9981. doi:10.1021/bi025907j. ISSN 0006-2960. PMID 12146961.
- ^ Kurreck, J. (2002-05-01). "Design of antisense oligonucleotides stabilized by locked nucleic acids". Nucleic Acids Research. 30 (9): 1911–1918. doi:10.1093/nar/30.9.1911. PMC 113840. PMID 11972327.
- ^ Frieden, M. (2003-11-01). "Expanding the design horizon of antisense oligonucleotides with alpha-L-LNA". Nucleic Acids Research. 31 (21): 6365–6372. doi:10.1093/nar/gkg820. ISSN 1362-4962. PMC 275462. PMID 14576324.
- ^ Frieden, Miriam; Hansen, Henrik F.; Koch, Troels (October 2003). "Nuclease Stability of LNA Oligonucleotides and LNA-DNA Chimeras". Nucleosides, Nucleotides and Nucleic Acids. 22 (5–8): 1041–1043. doi:10.1081/NCN-120022731. ISSN 1525-7770. PMID 14565339. S2CID 10631717.
- ^ Morita, K.; Hasegawa, C.; Kaneko, M.; Tsutsumi, S.; Sone, J.; Ishikawa, T.; Imanishi, T.; Koizumi, M. (2001-11-01). "2'-O, 4'-C-Ethylene-bridged nucleic acids (ENA) with nuclease-resistance and high affnity for RNA". Nucleic Acids Symposium Series. 1 (1): 241–242. doi:10.1093/nass/1.1.241. ISSN 0261-3166. PMID 12836354.
- ^ Veedu, Rakesh; Wengel, Jesper (2011). Medicinal Chemistry of Nucleic Acids. John Wiley & Sons, Inc. pp. 335–337. ISBN 978-0470596685.
- ^ a b Obika, Satoshi; Nanbu, Daishu; Hari, Yoshiyuki; Morio, Ken-ichiro; In, Yasuko; Ishida, Toshimasa; Imanishi, Takeshi (1997-12-15). "Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering". Tetrahedron Letters. 38 (50): 8735–8738. doi:10.1016/S0040-4039(97)10322-7. ISSN 0040-4039.
- ^ a b c Orum, Miriam Frieden and Henrik (2008-03-31). "Locked Nucleic Acid Holds Promise in the Treatment of Cancer". Current Pharmaceutical Design. 14 (11): 1138–1142. doi:10.2174/138161208784246234. PMID 18473860. Retrieved 2020-10-06.
- ^ Zamecnik, P. C.; Stephenson, M. L. (1978-01-01). "Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide". Proceedings of the National Academy of Sciences. 75 (1): 280–284. Bibcode:1978PNAS...75..280Z. doi:10.1073/pnas.75.1.280. ISSN 0027-8424. PMC 411230. PMID 75545.
- ^ Koshkin, Alexei A.; Fensholdt, Jef; Pfundheller, Henrik M.; Lomholt, Christian (2001-12-01). "A Simplified and Efficient Route to 2'-O, 4'-C-Methylene-Linked Bicyclic Ribonucleosides (Locked Nucleic Acid)". The Journal of Organic Chemistry. 66 (25): 8504–8512. doi:10.1021/jo010732p. ISSN 0022-3263. PMID 11735531.
- ^ a b Veedu, Rakesh N.; Vester, Birte; Wengel, Jesper (2007-03-26). "Enzymatic Incorporation of LNA Nucleotides into DNA Strands". ChemBioChem. 8 (5): 490–492. doi:10.1002/cbic.200600501. PMID 17315250. S2CID 10206060.
- ^ Breaker, R. R.; Joyce, G. F. (December 1994). "A DNA enzyme that cleaves RNA". Chemistry & Biology. 1 (4): 223–229. doi:10.1016/1074-5521(94)90014-0. ISSN 1074-5521. PMID 9383394.
- ^ Vester, Birte; Lundberg, Lars Bo; Sørensen, Mads D.; Babu, B. Ravindra; Douthwaite, Stephen; Wengel, Jesper (November 2002). "LNAzymes: Incorporation of LNA-Type Monomers into DNAzymes Markedly Increases RNA Cleavage". Journal of the American Chemical Society. 124 (46): 13682–13683. doi:10.1021/ja0276220. ISSN 0002-7863. PMID 12431091.
- ^ Schubert, Steffen; Fürste, Jens P; Werk, Denise; Grunert, Hans-Peter; Zeichhardt, Heinz; Erdmann, Volker A; Kurreck, Jens (May 2004). "Gaining Target Access for Deoxyribozymes". Journal of Molecular Biology. 339 (2): 355–363. doi:10.1016/j.jmb.2004.03.064. PMID 15136038.
- ^ a b Petersen M, Wengel J (February 2003). "LNA: a versatile tool for therapeutics and genomics". Trends in Biotechnology. 21 (2): 74–81. doi:10.1016/S0167-7799(02)00038-0. PMID 12573856.
- ^ Dürig, J.; Dührsen, U.; Klein-Hitpass, L.; Worm, J.; Hansen, J. B. Rode; Ørum, H.; Wissenbach, M. (April 2011). "The novel antisense Bcl-2 inhibitor SPC2996 causes rapid leukemic cell clearance and immune activation in chronic lymphocytic leukemia". Leukemia. 25 (4): 638–647. doi:10.1038/leu.2010.322. ISSN 1476-5551. PMID 21358717.
- ^ Gebert, Luca F. R.; Rebhan, Mario A. E.; Crivelli, Silvia E. M.; Denzler, Rémy; Stoffel, Markus; Hall, Jonathan (2014-01-01). "Miravirsen (SPC3649) can inhibit the biogenesis of miR-122". Nucleic Acids Research. 42 (1): 609–621. doi:10.1093/nar/gkt852. ISSN 0305-1048. PMC 3874169. PMID 24068553.
- ^ Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. (2019-06-24). "How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market". EJIFCC. 30 (2): 114–127. ISSN 1650-3414. PMC 6599191. PMID 31263388.
- ^ Bonetta L (2005). "Prime time for real-time PCR". Nat. Methods. 2 (4): 305–312. doi:10.1038/nmeth0405-305. S2CID 17711047.
- ^ a b c Kubota, Kengo; Ohashi, Akiyoshi; Imachi, Hiroyuki; Harada, Hideki (August 2006). "Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes". Applied and Environmental Microbiology. 72 (8): 5311–5317. doi:10.1128/AEM.03039-05. ISSN 0099-2240. PMC 1538721. PMID 16885281.
- ^ van Ravesteyn, TW; Dekker, M; Fish, A; Sixma, TK; Wolters, A; Dekker, RJ; Te Riele, HP (12 April 2016). "LNA modification of single-stranded DNA oligonucleotides allows subtle gene modification in mismatch-repair-proficient cells". Proceedings of the National Academy of Sciences of the United States of America. 113 (15): 4122–7. doi:10.1073/pnas.1513315113. PMC 4839440. PMID 26951689.