In molecular biology, zinc-dependent phospholipases C is a family of bacterial phospholipases C enzymes, some of which are also known as alpha toxins.
| Zinc dependent phospholipase C | |||||||||
|---|---|---|---|---|---|---|---|---|---|
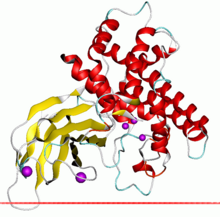 Alpha toxin of Clostridium showing the zinc dependent phospholipase domain in red and the PLAT domain in yellow | |||||||||
| Identifiers | |||||||||
| Symbol | Zn_dep_PLPC | ||||||||
| Pfam | PF00882 | ||||||||
| InterPro | IPR001531 | ||||||||
| PROSITE | PDOC00357 | ||||||||
| SCOP2 | 1ah7 / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 81 | ||||||||
| OPM protein | 1olp | ||||||||
| CDD | cd11009 | ||||||||
| |||||||||
Bacillus cereus contains a monomeric phospholipase C EC 3.1.4.3 (PLC) of 245 amino-acid residues. Although PLC prefers to act on phosphatidylcholine, it also shows weak catalytic activity with sphingomyelin and phosphatidylinositol.[1] Sequence studies have shown the protein to be similar both to alpha toxin from Clostridium perfringens and Clostridium bifermentans, a phospholipase C involved in haemolysis and cell rupture,[2] and to lecithinase from Listeria monocytogenes, which aids cell-to-cell spread by breaking down the 2-membrane vacuoles that surround the bacterium during transfer.[3]
Each of these proteins is a zinc-dependent enzyme, binding 3 zinc ions per molecule.[4] The enzymes catalyse the conversion of phosphatidylcholine and water to 1,2-diacylglycerol and choline phosphate.[1][2][4]
In Bacillus cereus, there are nine residues known to be involved in binding the zinc ions: 5 His, 2 Asp, 1 Glu and 1 Trp. These residues are all conserved in the Clostridium alpha-toxin.
Some examples of this enzyme contain a C-terminal sequence extension that contains a PLAT domain which is thought to be involved in membrane localisation.[5][6]
References
edit- ^ a b Nakamura S, Yamada A, Tsukagoshi N, Udaka S, Sasaki T, Makino S, Little C, Tomita M, Ikezawa H (1988). "Nucleotide sequence and expression in Escherichia coli of the gene coding for sphingomyelinase of Bacillus cereus". Eur. J. Biochem. 175 (2): 213–220. doi:10.1111/j.1432-1033.1988.tb14186.x. PMID 2841128.
- ^ a b Titball RW, Rubidge T, Hunter SE, Martin KL, Morris BC, Shuttleworth AD, Anderson DW, Kelly DC (1989). "Molecular cloning and nucleotide sequence of the alpha-toxin (phospholipase C) of Clostridium perfringens". Infect. Immun. 57 (2): 367–376. doi:10.1128/IAI.57.2.367-376.1989. PMC 313106. PMID 2536355.
- ^ Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P, Vazquez-Boland JA (1992). "Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread". Infect. Immun. 60 (1): 219–230. doi:10.1128/IAI.60.1.219-230.1992. PMC 257526. PMID 1309513.
- ^ a b Titball RW, Rubidge T (1990). "The role of histidine residues in the alpha toxin of Clostridium perfringens". FEMS Microbiol. Lett. 56 (3): 261–265. doi:10.1111/j.1574-6968.1988.tb03188.x. PMID 2111259.
- ^ Bateman A, Sandford R (1999). "The PLAT domain: a new piece in the PKD1 puzzle". Curr. Biol. 9 (16): R588–90. doi:10.1016/S0960-9822(99)80380-7. PMID 10469604. S2CID 15018010.
- ^ Ponting CP, Hofmann K, Bork P (August 1999). "A latrophilin/CL-1-like GPS domain in polycystin-1". Curr. Biol. 9 (16): R585–8. doi:10.1016/S0960-9822(99)80379-0. PMID 10469603. S2CID 17252179.