 | This is a user sandbox of Golgi101. You can use it for testing or practicing edits. This is not the sandbox where you should draft your assigned article for a dashboard.wikiedu.org course. To find the right sandbox for your assignment, visit your Dashboard course page and follow the Sandbox Draft link for your assigned article in the My Articles section. |
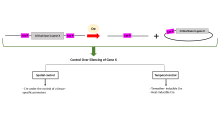
In genetics, floxing refers to the sandwiching of a piece of DNA sequence (which is then said to be floxed) between two lox P sites. The terms are constructed upon the phrase "flanking/flanked by lox P sites". Recombination between lox P sites is catalysed by Cre recombinase.[1] Depending on the placement of lox P sites, floxing a gene allows it to be deleted (knocked out),[2][3] inserted (knocked in),[4] translocated or inverted in a process called Cre-Lox recombination.
Floxed genes can be used to produce tissue-specific knockouts. For example, Cre recombinase under the control of alpha-myosin heavy chain[5] or Glial fibrillary acidic protein[6] promoter results in the floxed gene to be inactivated in the heart or astrocytes respectively. Further, these knockouts can be inducible. In several mouse studies, tamoxifen is used to induce the Cre recombinase.[7][8] In this case, Cre recombinase is fused to a portion of the mouse estrogen receptor (ERTM) which contains a mutation within its ligand binding domain (LBD). The mutation renders the receptor inactive, which leads to incorrect localization through its interactions with chaperone proteins such as heat shock protein 70 and 90 (Hsp70 and Hsp90). Tamoxifen binds to Cre-ERTM and disrupts its interactions with the chaperones, which allows the Cre-ERTM fusion protein to enter the nucleus and perform recombination on the floxed gene.[9][10] Additionally, Cre recombinase can be induced by heat when under the control of specific heat shock elements (HSEs).[11][12]
Relevance
editThe floxing of genes is essential in the development of scientific model systems as it allows the the expression of a gene to be manipulated in a specific tissue at any time chosen by the scientist. This means that researchers can have spatial and temporal control over gene expression. Moreover, animals such as mice can be used as models to study human disease. Therefore, Cre-lox system can be used in mice to manipulate gene expression in order to study human diseases and drug development.[13] For example, using the Cre-lox system, researchers can study Oncogenes and Tumor Supressor genes and their role in development and progression of cancer in mice models.[14]
References
edit- ^ Nagy A (2000). "Cre recombinase: the universal reagent for genome tailoring". Genesis. 26 (2): 99–109. doi:10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B. PMID 10686599.
- ^ Friedel, Roland H.; Wurst, Wolfgang; Wefers, Benedikt; Kühn, Ralf (2011). "Generating conditional knockout mice". Methods in Molecular Biology (Clifton, N.J.). 693: 205–231. doi:10.1007/978-1-60761-974-1_12. ISSN 1940-6029. PMID 21080282.
- ^ Sakamoto, Kazuhito; Gurumurthy, Channabasavaiah B.; Wagner, Kay-Uwe (2014), Singh, Shree Ram; Coppola, Vincenzo (eds.), "Generation of Conditional Knockout Mice", Mouse Genetics, vol. 1194, Springer New York, pp. 21–35, doi:10.1007/978-1-4939-1215-5_2, ISBN 9781493912148, retrieved 2019-10-07
- ^ Imuta, Yu; Kiyonari, Hiroshi; Jang, Chuan-Wei; Behringer, Richard R.; Sasaki, Hiroshi (2013). "Generation of knock-in mice that express nuclear enhanced green fluorescent protein and tamoxifen-inducible Cre recombinase in the notochord from Foxa2 and T loci: Mouse Lines Expressing EGFP and Inducible CRE". genesis. 51 (3): 210–218. doi:10.1002/dvg.22376. PMC 3632256. PMID 23359409.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Conditional mutagenesis : an approach to disease models. Abuin, A., Feil, Robert., Metzger, Daniel, Dr. Berlin: Springer. 2007. ISBN 9783540351092. OCLC 184984636.
{{cite book}}: CS1 maint: others (link) - ^ Astrocytes : methods and protocols. Milner, Richard, Professor,. New York: Human Press. 2012. ISBN 9781617794520. OCLC 769667324.
{{cite book}}: CS1 maint: extra punctuation (link) CS1 maint: others (link) - ^ Hayashi, Shigemi; McMahon, Andrew P. (2002). "Efficient Recombination in Diverse Tissues by a Tamoxifen-Inducible Form of Cre: A Tool for Temporally Regulated Gene Activation/Inactivation in the Mouse". Developmental Biology. 244 (2): 305–318. doi:10.1006/dbio.2002.0597.
- ^ Hayashi, Shigemi; McMahon, Andrew P. (2002). "Efficient Recombination in Diverse Tissues by a Tamoxifen-Inducible Form of Cre: A Tool for Temporally Regulated Gene Activation/Inactivation in the Mouse". Developmental Biology. 244 (2): 305–318. doi:10.1006/dbio.2002.0597. ISSN 0012-1606. PMID 11944939.
- ^ Danielian, P. S.; Muccino, D.; Rowitch, D. H.; Michael, S. K.; McMahon, A. P. (1998-12-03). "Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase". Current biology: CB. 8 (24): 1323–1326. doi:10.1016/s0960-9822(07)00562-3. ISSN 0960-9822. PMID 9843687.
- ^ Transgenesis techniques : principles and protocols. Clarke, Alan R. (2nd ed ed.). Totowa, NJ: Humana Press. 2002. ISBN 9781592591787. OCLC 50175106.
{{cite book}}:|edition=has extra text (help)CS1 maint: others (link) - ^ Cancer and zebrafish : mechanisms, techniques, and models. Langenau, David M.,. Switzerland. ISBN 9783319306544. OCLC 949668674.
{{cite book}}: CS1 maint: extra punctuation (link) CS1 maint: others (link) - ^ Kobayashi, Kayo; Kamei, Yasuhiro; Kinoshita, Masato; Czerny, Thomas; Tanaka, Minoru (2012). "A heat-inducible CRE/LOXP gene induction system in medaka". genesis. 51 (1): 59–67. doi:10.1002/dvg.22348.
- ^ Mouse genetics : methods and protocols. Singh, Shree Ram,, Coppola, Vincenzo,. New York, NY. ISBN 9781493912155. OCLC 885338722.
{{cite book}}: CS1 maint: extra punctuation (link) CS1 maint: others (link) - ^ "Genetically Engineered Mice for Cancer Research | SpringerLink" (PDF). doi:10.1007/978-0-387-69805-2.pdf.
{{cite journal}}: Cite journal requires|journal=(help)