Ferrichrome is a cyclic hexa-peptide that forms a complex with iron atoms. It is a siderophore composed of three glycine and three modified ornithine residues with hydroxamate groups [-N(OH)C(=O)C-]. The 6 oxygen atoms from the three hydroxamate groups bind Fe(III) in near perfect octahedral coordination.
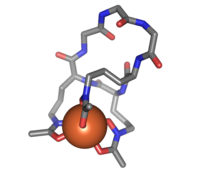 Ferrichrome (sticks) bound to an iron atom (orange)
| |
| Names | |
|---|---|
| IUPAC name
N-[3-[4,16-bis[3-[acetyl(oxido)amino]propyl]-2,5,8,11,14,17-hexaoxo-3,6,9,12,15,18-hexazacyclooctadec-1-yl]propyl]-N-oxidoacetamide; iron(3+)
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
| ECHA InfoCard | 100.036.081 |
| EC Number |
|
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C27H42FeN9O12 | |
| Molar mass | 740.529 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Ferrichrome was first isolated in 1952, and has been found to be produced by fungi of the genera Aspergillus, Ustilago, and Penicillium.[1] However, at the time there was no understanding regarding its involvement and contribution to iron transport.[2] It was not until 1957 because of Joe Neilands' work, where he first noted that Ferrichrome was able to act as an iron transport agent.
Biological function
editFerrichrome is a siderophore, which are metal chelating agents that have a low molecular mass and are produced by microorganisms and plants growing under low iron conditions. The main function of siderophores is to chelate ferric iron (Fe3+) from insoluble minerals from the environment and make it available for microbial and plant cells. Iron is important in biological functions as it acts as a catalyst in enzymatic processes, as well as for electron transfer, DNA and RNA synthesis, and oxygen metabolism.[3] Although iron is the fourth most abundant element in the Earth’s crust,[4] bioavailability of iron in aerobic environments is low due to formation of insoluble ferric hydroxides. Under iron limitation, bacteria scavenge for ferric iron (Fe3+) by up-regulating the secretion of siderophores in order to meet their nutritional requirements.[5] Recent studies have shown that ferrichrome has been used as a tumor- suppressive molecule produced by the bacterium Lacticaseibacillus casei. The study from the Department of Medicine and Asahikawa Medical University, suggests that ferrichrome has a greater tumor-suppressive effect than other drugs currently used to fight colon cancer, including cisplatin and 5-fluoro-uracil. Ferrichrome also had less of an effect on non-cancerous intestinal cells than the two previously mentioned cancer fighting drugs. It was determined that ferrichrome activated the C-Jun N-terminal kinases, which induced apoptosis. The induction of apoptosis by ferrichrome is reduced by the inhibition of the c-jun N-terminal kinase signaling pathway.[6]
Uptake
editIron is essential for the most important biological processes such as DNA and RNA synthesis, glycolysis, energy generation, nitrogen fixation and photosynthesis, therefore uptake of iron from the environment and transport into the organism are critical life processes for almost all organisms.[7] The problem is when environmental iron is exposed to oxygen it is mineralized to its insoluble ferric oxy hydroxide form which can not be transported into the cells and therefore is not available for use by the cell.[7] To overcome this, bacteria, fungi and some plants synthesize siderophores, and secrete it into an extracellular environment where binding of iron can occur.[7] It is important to note microbes make their own type of siderophore so that they are not competing with other organisms for iron uptake.[7] For example, saccharomyces cerevisiae is a species of yeast that can uptake the iron bound siderophore through transporters of the ARN family.[8] [Fe3+( siderophore)](n-3)- binds to a receptor-transporter on the cell surface and then is up taken.[8] The exact mechanism how iron enters the cell using these transporters is not understood, but it known that once it enters the cell it accumulates in the cytosol.[8] In saccharomyces cerevisiae, ferrichrome is specifically taken up by ARN1P as it has 2 binding sites and ferrichrome can the higher affinity site through endocytosis.[8] Ferrichrome chelates stay stable in the cell and allow for iron storage, but can be easily mobilized to meet the metabolic needs of the cell.[8]
The removal of Fe3+ occurs through the reduction of Fe3+ to Fe2+.[9] The reduction strategy helps in making the iron more aqueous soluble, and allows the iron to become more bioavailable in order for uptake to occur. This is because the Fe2+ product is not able to mineralize like the Fe3+, as it does not bind significantly to the chelate ligand that is designed to bind Fe3+. In addition to this, the Fe3+ product can also release Fe2+ from the chelate ligands that was designed to bind Fe3+. Fe2+ has little to no affinity towards the siderophore ligand and this removal is necessary for use and storage. This is because Fe2+ is an intermediate acid, therefore it is not able to bind significantly to the siderophore chelate ligands and can only bind with a much lower affinity. Whereas, Fe3+ is a hard base and can bind to the siderophore chelate ligands with a much higher affinity.[2] The Fe3+ siderophore complexes are taken up into the bacterial membrane by active transport mechanisms. This uptake process is able to recognize different structural features of the siderophores and transport the Fe3+ complexes into the periplasm.
Siderophore Binding
editThe main types of siderophores have catecholate, hydroxamate, and carboxylate coordinating ligands. An example of a catecholate siderophore includes enterobactin. Examples of hydroxamate siderophores include desferrioxamine, ferrichrome, aerobactin, rhodotorullic acid, and alcaligin. Aerobactin is a carboxylate siderophore as well. The triscatecholate siderophore, enterobactin, has a higher binding affinity of logβ110 = 49 to ferric iron compared to Ferrichrome, which has a binding affinity of logβ110 = 29.07. Therefore, it would outcompete with the other siderophore and bind more of the available environmental Fe3+. It does not bind other metals in high concentration because of its high Fe3+ specificity.[8] The trishydroxamate siderophore, desferrioxamine, has a binding affinity of logβ110 = 30.6 and has a lower binding affinity compared to Ferrichrome. Therefore, the desferrioxamine siderophore can also outcompete Ferrichrome, and bind more of the available environmental Fe3+. However, the bishydroxamate siderophores aerobactin (logβ110 = 22.5), rhodotorullic acid (logβ110 =21.55), and alcaligin (logβ110 = 23.5) will not be able to outcompete with the triscatecholate and trishydroxamate siderophores, since they do not have high Fe3+ specificity. Therefore, they are not able to bind more of the available environmental Fe3+.
Iron in its trivalent state has an electron configuration of d5, therefore, its complexes are preferentially hexacoordinate, quasi octahedral.[10] In terms of the HSAB principle, ferric siderophores have donor atoms that are mainly oxygen and rarely heterocyclic nitrogen. This is because of the ferric ion being a hard Lewis acid, and the ferric iron therefore binds more strongly with a hard anionic oxygen donor.
FhuA Uptake Mechanism
editE. coli has a receptor protein called FhuA (ferric Hydroxamate).[11]
FhuA’s is an energy-coupled transporter and receptor.[11] It is a part of the integral outer membrane proteins and works alongside an energy transducing protein TonB.[12] It is involved in the uptake of iron in complex with ferrichrome by binding and transporting ferrichrome-iron across the cell’s outer membrane.[12]
The green ribbons represent β-barrel wall that is 69Å long x 40-45Å diameter that represents the C-terminus residues. It has 22 antiparallel β strands. The blue ribbon in the center is a “cork” which is a distinct domain for the N-terminus residues.[12]
FhuA has L4 strand and its role is to transport ferrichrome into the β-barrel wall. The ferrichrome complex then binds tightly to both the β-barrel wall and the "cork".[12] As a result, these binding triggers two key conformation changes to iron-ferrichrome complex to transfer energy to the cork. This energy transfer results in subsequent conformational changes that transport iron-ferrichrome to the periplasmic pocket which signal a ligand loaded status of the receptor.[12] These subtle shifts disrupt the binding of iron-ferrichrome to the cork which then allows the permeation of the ferrichrome-iron to a putative channel-forming region. The inner wall of the β-barrel provides a series of weak binding sites to pull ferrichrome along.[12] FhuD is a high affinity binding protein in the periplasmic pocket that also aids in unidirectional transport across the cell envelope.[12]
See also
editReferences
edit- ^ Ferrichrome Archived 2010-01-13 at the Wayback Machine, Virtual Museum of Minerals and Molecules, University of Wisconsin
- ^ a b Kenneth Raymond - The Human/Bacterial Arms Race for Iron, retrieved 2021-12-04
- ^ Ahmed E, Holmström SJ (May 2014). "Siderophores in environmental research: roles and applications". Microbial Biotechnology. 7 (3): 196–208. doi:10.1111/1751-7915.12117. PMC 3992016. PMID 24576157.
- ^ Loper JE, Buyer JS (September 1990). "Siderophores in Microbial Interactions on Plant Surfaces". Molecular Plant-Microbe Interactions. 4: 5–13. doi:10.1094/mpmi-4-005.
- ^ Chatterjee A, O'Brian MR (April 2018). "Rapid evolution of a bacterial iron acquisition system". Molecular Microbiology. 108 (1): 90–100. doi:10.1111/mmi.13918. PMC 5867251. PMID 29381237.
- ^ Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J, et al. (August 2016). "Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis". Nature Communications. 7: 12365. doi:10.1038/ncomms12365. PMC 4987524. PMID 27507542.
- ^ a b c d Hannauer M, Barda Y, Mislin GL, Shanzer A, Schalk IJ (March 2010). "The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome". Journal of Bacteriology. 192 (5): 1212–1220. doi:10.1128/JB.01539-09. PMC 2820845. PMID 20047910.
- ^ a b c d e f Moore RE, Kim Y, Philpott CC (May 2003). "The mechanism of ferrichrome transport through Arn1p and its metabolism in Saccharomyces cerevisiae". Proceedings of the National Academy of Sciences of the United States of America. 100 (10): 5664–5669. Bibcode:2003PNAS..100.5664M. doi:10.1073/pnas.1030323100. PMC 156258. PMID 12721368.
- ^ Inomata T, Eguchi H, Funahashi Y, Ozawa T, Masuda H (January 2012). "Adsorption behavior of microbes on a QCM chip modified with an artificial siderophore-Fe3+ complex". Langmuir. 28 (2): 1611–1617. doi:10.1021/la203250n. PMID 22182317.
- ^ Drechsel H, Jung G (1998). "Peptide siderophores". Journal of Peptide Science. 4 (3): 147–181. doi:10.1002/(SICI)1099-1387(199805)4:3<147::AID-PSC136>3.0.CO;2-C. ISSN 1099-1387. PMID 9643626. S2CID 31107931.
- ^ a b Braun V (June 2009). "FhuA (TonA), the career of a protein". Journal of Bacteriology. 191 (11): 3431–3436. doi:10.1128/JB.00106-09. PMC 2681897. PMID 19329642.
- ^ a b c d e f g Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W (December 1998). "Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide". Science. 282 (5397): 2215–2220. Bibcode:1998Sci...282.2215F. doi:10.1126/science.282.5397.2215. PMID 9856937.