Aminolevulinic acid dehydratase (porphobilinogen synthase, or ALA dehydratase, or aminolevulinate dehydratase) is an enzyme (EC 4.2.1.24) that in humans is encoded by the ALAD gene.[5][6] Porphobilinogen synthase (or ALA dehydratase, or aminolevulinate dehydratase) synthesizes porphobilinogen through the asymmetric condensation of two molecules of aminolevulinic acid. All natural tetrapyrroles, including hemes, chlorophylls and vitamin B12, share porphobilinogen as a common precursor. Porphobilinogen synthase is the prototype morpheein.[7]
Function
editIt catalyzes the following reaction, the second step of the biosynthesis of porphyrin:
- 2 5-Aminolevulinic acid porphobilinogen + 2 H2O
It therefore catalyzes the condensation of 2 molecules of 5-aminolevulinate to form porphobilinogen (a precursor of heme, cytochromes and other hemoproteins). This reaction is the first common step in the biosynthesis of all biological tetrapyrroles. Zinc is essential for enzymatic activity.
Structure
editThe structural basis for allosteric regulation of Porphobilinogen synthase (PBGS) is modulation of a quaternary structure equilibrium between octamer and hexamer (via dimers), which is represented schematically as 6mer* ↔ 2mer* ↔ 2mer ↔ 8mer. The * represents a reorientation between two domains of each subunit that occurs in the dissociated state because it is sterically forbidden in the larger multimers.[7]
PBGS is encoded by a single gene and each PBGS multimer is composed of multiple copies of the same protein. Each PBGS subunit consists of a ~300 residue αβ-barrel domain, which houses the enzyme's active site in its center, and a >25 residue N-terminal arm domain. Allosteric regulation of PBGS can be described in terms of the orientation of the αβ-barrel domain with respect to the N-terminal arm domain.
Each N-terminal arm has up to two interactions with other subunits in a PBGS multimer. One of these interactions helps to stabilize a "closed" conformation of the active site lid. The other interaction restricts solvent access from the other end of the αβ-barrel.
In the inactive multimeric state, the N-terminal arm domain is not involved in the lid-stabilizing interaction, and in the crystal structure of the inactive assembly, the active site lid is disordered.
Allosteric regulators
editAs a nearly universal enzyme with a highly conserved active site, PBGS would not be a prime target for the development of antimicrobials and/or herbicides. To the contrary, allosteric sites can be much more phylogenetically variable than active sites, thus presenting more drug development opportunities.[7]
Phylogenetic variation in PBGS allostery leads to the framing of discussion of PBGS allosteric regulation in terms of intrinsic and extrinsic factors.
Intrinsic allosteric regulators
editMagnesium
editThe allosteric magnesium ion lies at the highly hydrated interface of two pro-octamer dimers. It appears to be easily dissociable, and it has been shown that hexamers accumulate when magnesium is removed in vitro.[8]
pH
editThough it is not common to consider hydronium ions as allosteric regulators, in the case of PBGS, side chain protonation at locations other than the active site has been shown to affect the quaternary structure equilibrium, and thus to affect the rate of its catalyzed reaction as well.
Extrinsic allosteric regulators
editSmall molecule hexamer stabilization
editInspection of the PBGS 6mer* reveals a surface cavity that is not present in the 8mer. Small molecule binding to this phylogenetically variable cavity has been proposed to stabilize 6mer* of the targeted PBGS and consequently inhibit activity.
Such allosteric regulators are known as morphlocks because they lock PBGS in a specific morpheein form (6mer*).[9]
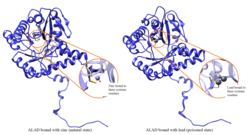
Lead poisoning
editALAD enzymatic activity is inhibited by lead, beginning at blood lead levels that were once considered to be safe (<10 μg/dL) and continuing to correlate negatively across the range from 5 to 95 μg/dL.[10] Inhibition of ALAD by lead leads to anemia primarily because it both inhibits heme synthesis and shortens the lifespan of circulating red blood cells, but also by stimulating the excessive production of the hormone erythropoietin, leading to inadequate maturation of red cells from their progenitors. A defect in the ALAD structural gene can cause increased sensitivity to lead poisoning and acute hepatic porphyria. Alternatively spliced transcript variants encoding different isoforms have been identified.[11]
Deficiency
editA deficiency of porphobilinogen synthase is usually acquired (rather than hereditary) and can be caused by heavy metal poisoning, especially lead poisoning, as the enzyme is very susceptible to inhibition by heavy metals.[12]
Hereditary insufficiency of porphobilinogen synthase is called porphobilinogen synthase (or ALA dehydratase) deficiency porphyria. It is an extremely rare cause of porphyria,[13] with less than 10 cases ever reported.[14] All disease associated protein variants favor hexamer formation relative to the wild type human enzyme.[13]
PBGS as the prototype morpheein
editThe morpheein model of allostery exemplified by PBGS adds an additional layer of understanding to potential mechanisms for regulation of protein function and complements the increased focus that the protein science community is placing on protein structure dynamics.[7]
This model illustrates how the dynamics of phenomena such as alternate protein conformations, alternate oligomeric states, and transient protein-protein interactions can be harnessed for allosteric regulation of catalytic activity.
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000148218 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000028393 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Eiberg H, Mohr J, Nielsen LS (February 1983). "delta-Aminolevulinatedehydrase: synteny with ABO-AK1-ORM (and assignment to chromosome 9)". Clinical Genetics. 23 (2): 150–4. doi:10.1111/j.1399-0004.1983.tb01864.x. PMID 6839527. S2CID 27267679.
- ^ Beaumont C, Foubert C, Grandchamp B, Weil D, Gross MS, Nordmann Y (May 1984). "Assignment of the human gene for delta aminolevulinate dehydrase to chromosome 9 by somatic cell hybridization and specific enzyme immunoassay". Annals of Human Genetics. 48 (2): 153–9. doi:10.1111/j.1469-1809.1984.tb01010.x. PMID 6378062. S2CID 24098976.
- ^ a b c d Jaffe EK, Lawrence SH (March 2012). "Allostery and the dynamic oligomerization of porphobilinogen synthase". Archives of Biochemistry and Biophysics. 519 (2): 144–53. doi:10.1016/j.abb.2011.10.010. PMC 3291741. PMID 22037356.
- ^ Breinig S, Kervinen J, Stith L, Wasson AS, Fairman R, Wlodawer A, et al. (September 2003). "Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase". Nature Structural Biology. 10 (9): 757–63. doi:10.1038/nsb963. PMID 12897770. S2CID 24188785.
- ^ Lawrence SH, Jaffe EK (2008). "Expanding the Concepts in Protein Structure-Function Relationships and Enzyme Kinetics: Teaching using Morpheeins". Biochemistry and Molecular Biology Education. 36 (4): 274–283. doi:10.1002/bmb.20211. PMC 2575429. PMID 19578473.
- ^ Abadin H, Ashizawa A, Stevens YW, Llados F, Diamond G, Sage G, Citra M, Quinones A, Bosch SJ, Swarts SG (August 2007). Toxicological Profile for Lead (PDF). Atlanta, GA: Agency for Toxic Substances and Disease Registry (US). pp. 22, 30. PMID 24049859. Retrieved 22 November 2015.
- ^ "Entrez Gene: ALAD aminolevulinate, delta-, dehydratase".
- ^ ALA dehydratase reaction, from NetBiochem at the University of Utah. Last modified 1/5/95
- ^ a b Jaffe EK, Stith L (February 2007). "ALAD porphyria is a conformational disease". American Journal of Human Genetics. 80 (2): 329–37. doi:10.1086/511444. PMC 1785348. PMID 17236137.
- ^ Overview of the Porphyrias Archived 2011-07-22 at the Wayback Machine at The Porphyrias Consortium (a part of NIH Rare Diseases Clinical Research Network (RDCRN)) Retrieved June 2011
External links
edit- Human ALAD genome location and ALAD gene details page in the UCSC Genome Browser.
- delta-Aminolevulinic+Acid+Dehydratase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- http://www.omim.org/entry/125270?search=pbgs&highlight=pbgs
Further reading
edit- Bernard A, Lauwerys R (1988). "Metal-induced alterations of delta-aminolevulinic acid dehydratase". Annals of the New York Academy of Sciences. 514: 41–7. doi:10.1111/j.1749-6632.1987.tb48759.x. PMID 3327436. S2CID 41966070.
- Jaffe EK (October 2004). "The porphobilinogen synthase catalyzed reaction mechanism". Bioorganic Chemistry. 32 (5): 316–25. doi:10.1016/j.bioorg.2004.05.010. PMID 15381398.
- Roels HA, Buchet JP, Lauwerys RR, Sonnet J (August 1975). "Comparison of in vivo effect of inorganic lead and cadmium on glutathione reductase system and delta-aminolevulinate dehydratase in human erythrocytes". British Journal of Industrial Medicine. 32 (3): 181–92. doi:10.1136/oem.32.3.181. PMC 1008057. PMID 1156566.
- Ishida N, Fujita H, Fukuda Y, Noguchi T, Doss M, Kappas A, Sassa S (May 1992). "Cloning and expression of the defective genes from a patient with delta-aminolevulinate dehydratase porphyria". The Journal of Clinical Investigation. 89 (5): 1431–7. doi:10.1172/JCI115732. PMC 443012. PMID 1569184.
- Dawson SJ, White LA (May 1992). "Treatment of Haemophilus aphrophilus endocarditis with ciprofloxacin". The Journal of Infection. 24 (3): 317–20. doi:10.1016/S0163-4453(05)80037-4. PMID 1602151.
- Astrin KH, Kaya AH, Wetmur JG, Desnick RJ (August 1991). "RsaI polymorphism in the human delta-aminolevulinate dehydratase gene at 9q34". Nucleic Acids Research. 19 (15): 4307. doi:10.1093/nar/19.15.4307-a. PMC 328595. PMID 1678509.
- Wetmur JG, Kaya AH, Plewinska M, Desnick RJ (October 1991). "Molecular characterization of the human delta-aminolevulinate dehydratase 2 (ALAD2) allele: implications for molecular screening of individuals for genetic susceptibility to lead poisoning". American Journal of Human Genetics. 49 (4): 757–63. PMC 1683158. PMID 1716854.
- Plewinska M, Thunell S, Holmberg L, Wetmur JG, Desnick RJ (July 1991). "delta-Aminolevulinate dehydratase deficient porphyria: identification of the molecular lesions in a severely affected homozygote". American Journal of Human Genetics. 49 (1): 167–74. PMC 1683193. PMID 2063868.
- Potluri VR, Astrin KH, Wetmur JG, Bishop DF, Desnick RJ (July 1987). "Human delta-aminolevulinate dehydratase: chromosomal localization to 9q34 by in situ hybridization". Human Genetics. 76 (3): 236–9. doi:10.1007/BF00283614. PMID 3036687. S2CID 32211471.
- Gibbs PN, Jordan PM (June 1986). "Identification of lysine at the active site of human 5-aminolaevulinate dehydratase". The Biochemical Journal. 236 (2): 447–51. doi:10.1042/bj2360447. PMC 1146860. PMID 3092810.
- Wetmur JG, Bishop DF, Cantelmo C, Desnick RJ (October 1986). "Human delta-aminolevulinate dehydratase: nucleotide sequence of a full-length cDNA clone". Proceedings of the National Academy of Sciences of the United States of America. 83 (20): 7703–7. Bibcode:1986PNAS...83.7703W. doi:10.1073/pnas.83.20.7703. PMC 386789. PMID 3463993.
- Wetmur JG, Bishop DF, Ostasiewicz L, Desnick RJ (1986). "Molecular cloning of a cDNA for human delta-aminolevulinate dehydratase". Gene. 43 (1–2): 123–30. doi:10.1016/0378-1119(86)90015-6. PMID 3758678.
- Doss M, von Tiepermann R, Schneider J (1981). "Acute hepatic porphyria syndrome with porphobilinogen synthase defect". The International Journal of Biochemistry. 12 (5–6): 823–6. doi:10.1016/0020-711X(80)90170-6. PMID 7450139.
- Kaya AH, Plewinska M, Wong DM, Desnick RJ, Wetmur JG (January 1994). "Human delta-aminolevulinate dehydratase (ALAD) gene: structure and alternative splicing of the erythroid and housekeeping mRNAs". Genomics. 19 (2): 242–8. doi:10.1006/geno.1994.1054. PMID 8188255.
- Akagi R, Yasui Y, Harper P, Sassa S (September 1999). "A novel mutation of delta-aminolaevulinate dehydratase in a healthy child with 12% erythrocyte enzyme activity". British Journal of Haematology. 106 (4): 931–7. doi:10.1046/j.1365-2141.1999.01647.x. PMID 10519994. S2CID 24044521.
- Akagi R, Shimizu R, Furuyama K, Doss MO, Sassa S (March 2000). "Novel molecular defects of the delta-aminolevulinate dehydratase gene in a patient with inherited acute hepatic porphyria". Hepatology. 31 (3): 704–8. doi:10.1002/hep.510310321. PMID 10706561. S2CID 8998084.
- Kervinen J, Jaffe EK, Stauffer F, Neier R, Wlodawer A, Zdanov A (July 2001). "Mechanistic basis for suicide inactivation of porphobilinogen synthase by 4,7-dioxosebacic acid, an inhibitor that shows dramatic species selectivity". Biochemistry. 40 (28): 8227–36. CiteSeerX 10.1.1.374.9639. doi:10.1021/bi010656k. PMID 11444968.