A bacteriophage (/bækˈtɪərioʊfeɪdʒ/), also known informally as a phage (/ˈfeɪdʒ/), is a virus that infects and replicates within bacteria and archaea. The term was derived from bacteria and the Ancient Greek word φαγεῖν (phagein), meaning 'to devour'. Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

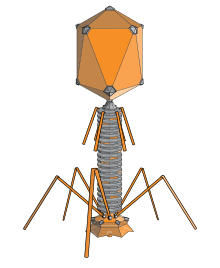

Bacteriophages are among the most common and diverse entities in the biosphere.[2] Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined.[3] Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biomass after prokaryotes,[4] where up to 9x108 virions per millilitre have been found in microbial mats at the surface,[5] and up to 70% of marine bacteria may be infected by bacteriophages.[6]
Bacteriophages were used from the 1920s as an alternative to antibiotics in the former Soviet Union and Central Europe, as well as in France.[7][8] They are seen as a possible therapy against multi-drug-resistant strains of many bacteria (see phage therapy).[9][10][11][12]
Bacteriophages are known to interact with the immune system both indirectly via bacterial expression of phage-encoded proteins and directly by influencing innate immunity and bacterial clearance.[13] Phage–host interactions are becoming increasingly important areas of research.[14]
Classification
editBacteriophages occur abundantly in the biosphere, with different genomes and lifestyles. Phages are classified by the International Committee on Taxonomy of Viruses (ICTV) according to morphology and nucleic acid.
| Order | Family | Morphology | Nucleic acid | Examples |
|---|---|---|---|---|
| Belfryvirales | Turriviridae | Enveloped, isometric | Linear dsDNA | |
| Caudovirales | Ackermannviridae | Nonenveloped, contractile tail | Linear dsDNA | |
| Autographiviridae | Nonenveloped, noncontractile tail (short) | Linear dsDNA | ||
| Chaseviridae | Linear dsDNA | |||
| Demerecviridae | Linear dsDNA | |||
| Drexlerviridae | Linear dsDNA | |||
| Guenliviridae | Linear dsDNA | |||
| Herelleviridae | Nonenveloped, contractile tail | Linear dsDNA | ||
| Myoviridae | Nonenveloped, contractile tail | Linear dsDNA | T4, Mu, P1, P2 | |
| Siphoviridae | Nonenveloped, noncontractile tail (long) | Linear dsDNA | λ, T5, HK97, N15 | |
| Podoviridae | Nonenveloped, noncontractile tail (short) | Linear dsDNA | T7, T3, Φ29, P22 | |
| Rountreeviridae | Linear dsDNA | |||
| Salasmaviridae | Linear dsDNA | |||
| Schitoviridae | Linear dsDNA | |||
| Zobellviridae | Linear dsDNA | |||
| Halopanivirales | Sphaerolipoviridae | Enveloped, isometric | Linear dsDNA | |
| Simuloviridae | Enveloped, isometric | Linear dsDNA | ||
| Matshushitaviridae | Enveloped, isometric | Linear dsDNA | ||
| Haloruvirales | Pleolipoviridae | Enveloped, pleomorphic | Circular ssDNA, circular dsDNA, or linear dsDNA | |
| Kalamavirales | Tectiviridae | Nonenveloped, isometric | Linear dsDNA | |
| Ligamenvirales | Lipothrixviridae | Enveloped, rod-shaped | Linear dsDNA | Acidianus filamentous virus 1 |
| Rudiviridae | Nonenveloped, rod-shaped | Linear dsDNA | Sulfolobus islandicus rod-shaped virus 1 | |
| Mindivirales | Cystoviridae | Enveloped, spherical | Linear dsRNA | Φ6 |
| Norzivirales | Atkinsviridae | Nonenveloped, isometric | Linear ssRNA | |
| Duinviridae | Nonenveloped, isometric | Linear ssRNA | ||
| Fiersviridae | Nonenveloped, isometric | Linear ssRNA | MS2, Qβ | |
| Solspiviridae | Nonenveloped, isometric | Linear ssRNA | ||
| Petitvirales | Microviridae | Nonenveloped, isometric | Circular ssDNA | ΦX174 |
| Primavirales | Tristromaviridae | Enveloped, rod-shaped | Linear dsDNA | |
| Timlovirales | Blumeviridae | Nonenveloped, isometric | Linear ssRNA | |
| Steitzviridae | Nonenveloped, isometric | Linear ssRNA | ||
| Tubulavirales | Inoviridae | Nonenveloped, filamentous | Circular ssDNA | M13 |
| Paulinoviridae | Nonenveloped, filamentous | Circular ssDNA | ||
| Plectroviridae | Nonenveloped, filamentous | Circular ssDNA | ||
| Vinavirales | Corticoviridae | Nonenveloped, isometric | Circular dsDNA | PM2 |
| Durnavirales | Picobirnaviridae (proposal) | Nonenveloped, isometric | Linear dsRNA | |
| Unassigned | Ampullaviridae | Enveloped, bottle-shaped | Linear dsDNA | |
| Autolykiviridae | Nonenveloped, isometric | Linear dsDNA | ||
| Bicaudaviridae | Nonenveloped, lemon-shaped | Circular dsDNA | ||
| Clavaviridae | Nonenveloped, rod-shaped | Circular dsDNA | ||
| Finnlakeviridae | Nonenveloped, isometric | Circular ssDNA | FLiP[15] | |
| Fuselloviridae | Nonenveloped, lemon-shaped | Circular dsDNA | Alphafusellovirus | |
| Globuloviridae | Enveloped, isometric | Linear dsDNA | ||
| Guttaviridae | Nonenveloped, ovoid | Circular dsDNA | ||
| Halspiviridae | Nonenveloped, lemon-shaped | Linear dsDNA | ||
| Plasmaviridae | Enveloped, pleomorphic | Circular dsDNA | ||
| Portogloboviridae | Enveloped, isometric | Circular dsDNA | ||
| Thaspiviridae | Nonenveloped, lemon-shaped | Linear dsDNA | ||
| Spiraviridae | Nonenveloped, rod-shaped | Circular ssDNA |
It has been suggested that members of Picobirnaviridae infect bacteria, but not mammals.[16]
There are also many unassigned genera of the class Leviviricetes: Chimpavirus, Hohglivirus, Mahrahvirus, Meihzavirus, Nicedsevirus, Sculuvirus, Skrubnovirus, Tetipavirus and Winunavirus containing linear ssRNA genomes[17] and the unassigned genus Lilyvirus of the order Caudovirales containing a linear dsDNA genome.
History
editIn 1896, Ernest Hanbury Hankin reported that something in the waters of the Ganges and Yamuna rivers in India had a marked antibacterial action against cholera and it could pass through a very fine porcelain filter.[18] In 1915, British bacteriologist Frederick Twort, superintendent of the Brown Institution of London, discovered a small agent that infected and killed bacteria. He believed the agent must be one of the following:
- a stage in the life cycle of the bacteria
- an enzyme produced by the bacteria themselves, or
- a virus that grew on and destroyed the bacteria[19]
Twort's research was interrupted by the onset of World War I, as well as a shortage of funding and the discoveries of antibiotics.
Independently, French-Canadian microbiologist Félix d'Hérelle, working at the Pasteur Institute in Paris, announced on 3 September 1917 that he had discovered "an invisible, antagonistic microbe of the dysentery bacillus". For d'Hérelle, there was no question as to the nature of his discovery: "In a flash I had understood: what caused my clear spots was in fact an invisible microbe... a virus parasitic on bacteria."[20] D'Hérelle called the virus a bacteriophage, a bacterium-eater (from the Greek phagein, meaning "to devour"). He also recorded a dramatic account of a man suffering from dysentery who was restored to good health by the bacteriophages.[21] It was d'Hérelle who conducted much research into bacteriophages and introduced the concept of phage therapy.[22] In 1919, in Paris, France, d'Hérelle conducted the first clinical application of a bacteriophage, with the first reported use in the United States being in 1922.[23]
Nobel prizes awarded for phage research
editIn 1969, Max Delbrück, Alfred Hershey, and Salvador Luria were awarded the Nobel Prize in Physiology or Medicine for their discoveries of the replication of viruses and their genetic structure.[24] Specifically the work of Hershey, as contributor to the Hershey–Chase experiment in 1952, provided convincing evidence that DNA, not protein, was the genetic material of life. Delbrück and Luria carried out the Luria–Delbrück experiment which demonstrated statistically that mutations in bacteria occur randomly and thus follow Darwinian rather than Lamarckian principles.
Uses
editPhage therapy
editPhages were discovered to be antibacterial agents and were used in the former Soviet Republic of Georgia (pioneered there by Giorgi Eliava with help from the co-discoverer of bacteriophages, Félix d'Hérelle) during the 1920s and 1930s for treating bacterial infections.
D'Herelle "quickly learned that bacteriophages are found wherever bacteria thrive: in sewers, in rivers that catch waste runoff from pipes, and in the stools of convalescent patients."[25]
They had widespread use, including treatment of soldiers in the Red Army.[26] However, they were abandoned for general use in the West for several reasons:
- Antibiotics were discovered and marketed widely. They were easier to make, store, and prescribe.
- Medical trials of phages were carried out, but a basic lack of understanding of phages raised questions about the validity of these trials.[27]
- Publication of research in the Soviet Union was mainly in the Russian or Georgian languages and for many years was not followed internationally.
- The Soviet technology was widely discouraged and in some cases illegal due to the red scare.
The use of phages has continued since the end of the Cold War in Russia,[28] Georgia, and elsewhere in Central and Eastern Europe. The first regulated, randomized, double-blind clinical trial was reported in the Journal of Wound Care in June 2009, which evaluated the safety and efficacy of a bacteriophage cocktail to treat infected venous ulcers of the leg in human patients.[29] The FDA approved the study as a Phase I clinical trial. The study's results demonstrated the safety of therapeutic application of bacteriophages, but did not show efficacy. The authors explained that the use of certain chemicals that are part of standard wound care (e.g. lactoferrin or silver) may have interfered with bacteriophage viability.[29] Shortly after that, another controlled clinical trial in Western Europe (treatment of ear infections caused by Pseudomonas aeruginosa) was reported in the journal Clinical Otolaryngology in August 2009.[30] The study concludes that bacteriophage preparations were safe and effective for treatment of chronic ear infections in humans. Additionally, there have been numerous animal and other experimental clinical trials evaluating the efficacy of bacteriophages for various diseases, such as infected burns and wounds, and cystic fibrosis-associated lung infections, among others.[30] On the other hand, phages of Inoviridae have been shown to complicate biofilms involved in pneumonia and cystic fibrosis and to shelter the bacteria from drugs meant to eradicate disease, thus promoting persistent infection.[31]
Meanwhile, bacteriophage researchers have been developing engineered viruses to overcome antibiotic resistance, and engineering the phage genes responsible for coding enzymes that degrade the biofilm matrix, phage structural proteins, and the enzymes responsible for lysis of the bacterial cell wall.[5][6][7] There have been results showing that T4 phages that are small in size and short-tailed can be helpful in detecting E. coli in the human body.[32]
Therapeutic efficacy of a phage cocktail was evaluated in a mouse model with nasal infection of multi-drug-resistant (MDR) A. baumannii. Mice treated with the phage cocktail showed a 2.3-fold higher survival rate compared to those untreated at seven days post-infection.[33]
In 2017, a 68-year-old diabetic patient with necrotizing pancreatitis complicated by a pseudocyst infected with MDR A. baumannii strains was being treated with a cocktail of Azithromycin, Rifampicin, and Colistin for 4 months without results and overall rapidly declining health.
Because discussion had begun of the clinical futility of further treatment, an Emergency Investigational New Drug (eIND) was filed as a last effort to at the very least gain valuable medical data from the situation, and approved, so he was subjected to phage therapy using a percutaneously (PC) injected cocktail containing nine different phages that had been identified as effective against the primary infection strain by rapid isolation and testing techniques (a process which took under a day). This proved effective for a very brief period, although the patient remained unresponsive and his health continued to worsen; soon isolates of a strain of A. baumannii were being collected from drainage of the cyst that showed resistance to this cocktail, and a second cocktail which was tested to be effective against this new strain was added, this time by intravenous (IV) injection as it had become clear that the infection was more pervasive than originally thought.[34]
Once on the combination of the IV and PC therapy the patient's downward clinical trajectory reversed, and within two days he had awoken from his coma and become responsive. As his immune system began to function he had to be temporarily removed from the cocktail because his fever was spiking to over 104 °F (40 °C), but after two days the phage cocktails were re-introduced at levels he was able to tolerate. The original three-antibiotic cocktail was replaced by minocycline after the bacterial strain was found not to be resistant to this and he rapidly regained full lucidity, although he was not discharged from the hospital until roughly 145 days after phage therapy began. Towards the end of the therapy it was discovered that the bacteria had become resistant to both of the original phage cocktails, but they were continued because they seemed to be preventing minocycline resistance from developing in the bacterial samples collected so were having a useful synergistic effect.[34]
Other
editFood industry
editPhages have increasingly been used to safen food products and to forestall spoilage bacteria.[35] Since 2006, the United States Food and Drug Administration (FDA) and United States Department of Agriculture (USDA) have approved several bacteriophage products. LMP-102 (Intralytix) was approved for treating ready-to-eat (RTE) poultry and meat products. In that same year, the FDA approved LISTEX (developed and produced by Micreos) using bacteriophages on cheese to kill Listeria monocytogenes bacteria, in order to give them generally recognized as safe (GRAS) status.[36] In July 2007, the same bacteriophage were approved for use on all food products.[37] In 2011 USDA confirmed that LISTEX is a clean label processing aid and is included in USDA.[38] Research in the field of food safety is continuing to see if lytic phages are a viable option to control other food-borne pathogens in various food products.[39]
Water indicators
editBacteriophages, including those specific to Escherichia coli, have been employed as indicators of fecal contamination in water sources. Due to their shared structural and biological characteristics, coliphages can serve as proxies for viral fecal contamination and the presence of pathogenic viruses such as rotavirus, norovirus, and HAV. Research conducted on wastewater treatment systems has revealed significant disparities in the behavior of coliphages compared to fecal coliforms, demonstrating a distinct correlation with the recovery of pathogenic viruses at the treatment's conclusion. Establishing a secure discharge threshold, studies have determined that discharges below 3000 PFU/100 mL are considered safe in terms of limiting the release of pathogenic viruses.[40]
Diagnostics
editIn 2011, the FDA cleared the first bacteriophage-based product for in vitro diagnostic use.[41] The KeyPath MRSA/MSSA Blood Culture Test uses a cocktail of bacteriophage to detect Staphylococcus aureus in positive blood cultures and determine methicillin resistance or susceptibility. The test returns results in about five hours, compared to two to three days for standard microbial identification and susceptibility test methods. It was the first accelerated antibiotic-susceptibility test approved by the FDA.[42]
Counteracting bioweapons and toxins
editGovernment agencies in the West have for several years been looking to Georgia and the former Soviet Union for help with exploiting phages for counteracting bioweapons and toxins, such as anthrax and botulism.[43] Developments are continuing among research groups in the U.S. Other uses include spray application in horticulture for protecting plants and vegetable produce from decay and the spread of bacterial disease. Other applications for bacteriophages are as biocides for environmental surfaces, e.g., in hospitals, and as preventative treatments for catheters and medical devices before use in clinical settings. The technology for phages to be applied to dry surfaces, e.g., uniforms, curtains, or even sutures for surgery now exists. Clinical trials reported in Clinical Otolaryngology[30] show success in veterinary treatment of pet dogs with otitis.
Bacterium sensing and identification
editThe sensing of phage-triggered ion cascades (SEPTIC) bacterium sensing and identification method uses the ion emission and its dynamics during phage infection and offers high specificity and speed for detection.[44]
Phage display
editPhage display is a different use of phages involving a library of phages with a variable peptide linked to a surface protein. Each phage genome encodes the variant of the protein displayed on its surface (hence the name), providing a link between the peptide variant and its encoding gene. Variant phages from the library may be selected through their binding affinity to an immobilized molecule (e.g., botulism toxin) to neutralize it. The bound, selected phages can be multiplied by reinfecting a susceptible bacterial strain, thus allowing them to retrieve the peptides encoded in them for further study.[45]
Antimicrobial drug discovery
editPhage proteins often have antimicrobial activity and may serve as leads for peptidomimetics, i.e. drugs that mimic peptides.[46] Phage-ligand technology makes use of phage proteins for various applications, such as binding of bacteria and bacterial components (e.g. endotoxin) and lysis of bacteria.[47]
Basic research
editBacteriophages are important model organisms for studying principles of evolution and ecology.[48]
Detriments
editDairy industry
editBacteriophages present in the environment can cause cheese to not ferment. In order to avoid this, mixed-strain starter cultures and culture rotation regimes can be used.[49] Genetic engineering of culture microbes – especially Lactococcus lactis and Streptococcus thermophilus – have been studied for genetic analysis and modification to improve phage resistance. This has especially focused on plasmid and recombinant chromosomal modifications.[50][35]
Some research has focused on the potential of bacteriophages as antimicrobial against foodborne pathogens and biofilm formation within the dairy industry. As the spread of antibiotic resistance is a main concern within the dairy industry, phages can serve as a promising alternative.[51]
Replication
editThe life cycle of bacteriophages tends to be either a lytic cycle or a lysogenic cycle. In addition, some phages display pseudolysogenic behaviors.[13]
With lytic phages such as the T4 phage, bacterial cells are broken open (lysed) and destroyed after immediate replication of the virion. As soon as the cell is destroyed, the phage progeny can find new hosts to infect.[13] Lytic phages are more suitable for phage therapy. Some lytic phages undergo a phenomenon known as lysis inhibition, where completed phage progeny will not immediately lyse out of the cell if extracellular phage concentrations are high. This mechanism is not identical to that of the temperate phage going dormant and usually is temporary.[52]
In contrast, the lysogenic cycle does not result in immediate lysing of the host cell. Those phages able to undergo lysogeny are known as temperate phages. Their viral genome will integrate with host DNA and replicate along with it, relatively harmlessly, or may even become established as a plasmid. The virus remains dormant until host conditions deteriorate, perhaps due to depletion of nutrients, then, the endogenous phages (known as prophages) become active. At this point they initiate the reproductive cycle, resulting in lysis of the host cell. As the lysogenic cycle allows the host cell to continue to survive and reproduce, the virus is replicated in all offspring of the cell. An example of a bacteriophage known to follow the lysogenic cycle and the lytic cycle is the phage lambda of E. coli.[53]
Sometimes prophages may provide benefits to the host bacterium while they are dormant by adding new functions to the bacterial genome, in a phenomenon called lysogenic conversion. Examples are the conversion of harmless strains of Corynebacterium diphtheriae or Vibrio cholerae by bacteriophages to highly virulent ones that cause diphtheria or cholera, respectively.[54][55] Strategies to combat certain bacterial infections by targeting these toxin-encoding prophages have been proposed.[56]
Attachment and penetration
editBacterial cells are protected by a cell wall of polysaccharides, which are important virulence factors protecting bacterial cells against both immune host defenses and antibiotics.[57] To enter a host cell, bacteriophages bind to specific receptors on the surface of bacteria, including lipopolysaccharides, teichoic acids, proteins, or even flagella. This specificity means a bacteriophage can infect only certain bacteria bearing receptors to which they can bind, which in turn, determines the phage's host range. Polysaccharide-degrading enzymes are virion-associated proteins that enzymatically degrade the capsular outer layer of their hosts at the initial step of a tightly programmed phage infection process.[citation needed] Host growth conditions also influence the ability of the phage to attach and invade them.[58] As phage virions do not move independently, they must rely on random encounters with the correct receptors when in solution, such as blood, lymphatic circulation, irrigation, soil water, etc.[citation needed]
Myovirus bacteriophages use a hypodermic syringe-like motion to inject their genetic material into the cell. After contacting the appropriate receptor, the tail fibers flex to bring the base plate closer to the surface of the cell. This is known as reversible binding. Once attached completely, irreversible binding is initiated and the tail contracts, possibly with the help of ATP present in the tail,[6] injecting genetic material through the bacterial membrane.[59] The injection is accomplished through a sort of bending motion in the shaft by going to the side, contracting closer to the cell and pushing back up. Podoviruses lack an elongated tail sheath like that of a myovirus, so instead, they use their small, tooth-like tail fibers enzymatically to degrade a portion of the cell membrane before inserting their genetic material.
Synthesis of proteins and nucleic acid
editWithin minutes, bacterial ribosomes start translating viral mRNA into protein. For RNA-based phages, RNA replicase is synthesized early in the process. Proteins modify the bacterial RNA polymerase so it preferentially transcribes viral mRNA. The host's normal synthesis of proteins and nucleic acids is disrupted, and it is forced to manufacture viral products instead. These products go on to become part of new virions within the cell, helper proteins that contribute to the assemblage of new virions, or proteins involved in cell lysis. In 1972, Walter Fiers (University of Ghent, Belgium) was the first to establish the complete nucleotide sequence of a gene and in 1976, of the viral genome of bacteriophage MS2.[60] Some dsDNA bacteriophages encode ribosomal proteins, which are thought to modulate protein translation during phage infection.[61]
Virion assembly
editIn the case of the T4 phage, the construction of new virus particles involves the assistance of helper proteins that act catalytically during phage morphogenesis.[62] The base plates are assembled first, with the tails being built upon them afterward. The head capsids, constructed separately, will spontaneously assemble with the tails. During assembly of the phage T4 virion, the morphogenetic proteins encoded by the phage genes interact with each other in a characteristic sequence. Maintaining an appropriate balance in the amounts of each of these proteins produced during viral infection appears to be critical for normal phage T4 morphogenesis.[63] The DNA is packed efficiently within the heads.[64] The whole process takes about 15 minutes.
Early studies of bactioriophage T4 (1962-1964) provided an opportunity to gain understanding of virtually all of the genes that are essential for growth of the bacteriophage under laboratory conditions.[65][66] These studies were made possible by the availability of two classes of conditional lethal mutants.[67] One class of such mutants was referred to as amber mutants.[67] The other class of conditional lethal mutants was referred to as temperature-sensitive mutants[68] Studies of these two classes of mutants led to considerable insight into the functions and interactions of the proteins employed in the machinery of DNA replication, repair and recombination, and on how viruses are assembled from protein and nucleic acid components (molecular morphogenesis).
Release of virions
editPhages may be released via cell lysis, by extrusion, or, in a few cases, by budding. Lysis, by tailed phages, is achieved by an enzyme called endolysin, which attacks and breaks down the cell wall peptidoglycan. An altogether different phage type, the filamentous phage, makes the host cell continually secrete new virus particles. Released virions are described as free, and, unless defective, are capable of infecting a new bacterium. Budding is associated with certain Mycoplasma phages. In contrast to virion release, phages displaying a lysogenic cycle do not kill the host and instead become long-term residents as prophages.[69]
Communication
editResearch in 2017 revealed that the bacteriophage Φ3T makes a short viral protein that signals other bacteriophages to lie dormant instead of killing the host bacterium. Arbitrium is the name given to this protein by the researchers who discovered it.[70][71]
Genome structure
editGiven the millions of different phages in the environment, phage genomes come in a variety of forms and sizes. RNA phages such as MS2 have the smallest genomes, with only a few kilobases. However, some DNA phages such as T4 may have large genomes with hundreds of genes; the size and shape of the capsid varies along with the size of the genome.[72] The largest bacteriophage genomes reach a size of 735 kb.[73]
Bacteriophage genomes can be highly mosaic, i.e. the genome of many phage species appear to be composed of numerous individual modules. These modules may be found in other phage species in different arrangements. Mycobacteriophages, bacteriophages with mycobacterial hosts, have provided excellent examples of this mosaicism. In these mycobacteriophages, genetic assortment may be the result of repeated instances of site-specific recombination and illegitimate recombination (the result of phage genome acquisition of bacterial host genetic sequences).[75] Evolutionary mechanisms shaping the genomes of bacterial viruses vary between different families and depend upon the type of the nucleic acid, characteristics of the virion structure, as well as the mode of the viral life cycle.[76]
Some marine roseobacter phages contain deoxyuridine (dU) instead of deoxythymidine (dT) in their genomic DNA. There is some evidence that this unusual component is a mechanism to evade bacterial defense mechanisms such as restriction endonucleases and CRISPR/Cas systems which evolved to recognize and cleave sequences within invading phages, thereby inactivating them. Other phages have long been known to use unusual nucleotides. In 1963, Takahashi and Marmur identified a Bacillus phage that has dU substituting dT in its genome,[77] and in 1977, Kirnos et al. identified a cyanophage containing 2-aminoadenine (Z) instead of adenine (A).[78]
Systems biology
editThe field of systems biology investigates the complex networks of interactions within an organism, usually using computational tools and modeling.[79] For example, a phage genome that enters into a bacterial host cell may express hundreds of phage proteins which will affect the expression of numerous host genes or the host's metabolism. All of these complex interactions can be described and simulated in computer models.[79]
For instance, infection of Pseudomonas aeruginosa by the temperate phage PaP3 changed the expression of 38% (2160/5633) of its host's genes. Many of these effects are probably indirect, hence the challenge becomes to identify the direct interactions among bacteria and phage.[80]
Several attempts have been made to map protein–protein interactions among phage and their host. For instance, bacteriophage lambda was found to interact with its host, E. coli, by dozens of interactions. Again, the significance of many of these interactions remains unclear, but these studies suggest that there most likely are several key interactions and many indirect interactions whose role remains uncharacterized.[81]
Host resistance
editBacteriophages are a major threat to bacteria and prokaryotes have evolved numerous mechanisms to block infection or to block the replication of bacteriophages within host cells. The CRISPR system is one such mechanism as are retrons and the anti-toxin system encoded by them.[82] The Thoeris defense system is known to deploy a unique strategy for bacterial antiphage resistance via NAD+ degradation.[83]
Bacteriophage–host symbiosis
editTemperate phages are bacteriophages that integrate their genetic material into the host as extrachromosomal episomes or as a prophage during a lysogenic cycle.[84][85][86] Some temperate phages can confer fitness advantages to their host in numerous ways, including giving antibiotic resistance through the transfer or introduction of antibiotic resistance genes (ARGs),[85][87] protecting hosts from phagocytosis,[88][89] protecting hosts from secondary infection through superinfection exclusion,[90][91][92] enhancing host pathogenicity,[84][93] or enhancing bacterial metabolism or growth.[94][95][96][97] Bacteriophage–host symbiosis may benefit bacteria by providing selective advantages while passively replicating the phage genome.[98]
In the environment
editMetagenomics has allowed the in-water detection of bacteriophages that was not possible previously.[99]
Also, bacteriophages have been used in hydrological tracing and modelling in river systems, especially where surface water and groundwater interactions occur. The use of phages is preferred to the more conventional dye marker because they are significantly less absorbed when passing through ground waters and they are readily detected at very low concentrations.[100] Non-polluted water may contain approximately 2×108 bacteriophages per ml.[101]
Bacteriophages are thought to contribute extensively to horizontal gene transfer in natural environments, principally via transduction, but also via transformation.[102] Metagenomics-based studies also have revealed that viromes from a variety of environments harbor antibiotic-resistance genes, including those that could confer multidrug resistance.[103]
Recent findings have mapped the complex and intertwined arsenal of anti-phage defense tools in environmental bacteria.[104]
In humans
editAlthough phages do not infect humans, there are countless phage particles in the human body, given the extensive human microbiome. One's phage population has been called the human phageome, including the "healthy gut phageome" (HGP) and the "diseased human phageome" (DHP).[105] The active phageome of a healthy human (i.e., actively replicating as opposed to nonreplicating, integrated prophage) has been estimated to comprise dozens to thousands of different viruses.[106] There is evidence that bacteriophages and bacteria interact in the human gut microbiome both antagonistically and beneficially.[107]
Preliminary studies have indicated that common bacteriophages are found in 62% of healthy individuals on average, while their prevalence was reduced by 42% and 54% on average in patients with ulcerative colitis (UC) and Crohn's disease (CD).[105] Abundance of phages may also decline in the elderly.[107]
The most common phages in the human intestine, found worldwide, are crAssphages. CrAssphages are transmitted from mother to child soon after birth, and there is some evidence suggesting that they may be transmitted locally. Each person develops their own unique crAssphage clusters. CrAss-like phages also may be present in primates besides humans.[107]
Commonly studied bacteriophages
editAmong the countless phages, only a few have been studied in detail, including some historically important phage that were discovered in the early days of microbial genetics. These, especially the T-phage, helped to discover important principles of gene structure and function.
Bacteriophage databases and resources
editSee also
edit- Antibiotic
- Bacterivore
- CrAssphage
- CRISPR
- DNA viruses
- Macrophage
- Phage ecology
- Phage monographs (a comprehensive listing of phage and phage-associated monographs, 1921–present)
- Phagemid
- Polyphage
- RNA viruses
- Transduction
- Viriome
- Virophage, viruses that infect other viruses
References
edit- ^ Padilla-Sanchez V (2021). "Structural Model of Bacteriophage T4". WikiJournal of Science. 4 (1): 5. doi:10.15347/WJS/2021.005. S2CID 238939621.
- ^ a b McGrath S, van Sinderen D, eds. (2007). Bacteriophage: Genetics and Molecular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-14-1.
- ^ LaFee S, Buschman H (25 April 2017). "Novel Phage Therapy Saves Patient with Multidrug-Resistant Bacterial Infection". UC Health – UC San Diego. Retrieved 13 May 2018.
- ^ Suttle CA (September 2005). "Viruses in the sea". Nature. 437 (7057): 356–361. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. PMID 16163346. S2CID 4370363.
- ^ a b Wommack KE, Colwell RR (March 2000). "Virioplankton: viruses in aquatic ecosystems". Microbiology and Molecular Biology Reviews. 64 (1): 69–114. doi:10.1128/MMBR.64.1.69-114.2000. PMC 98987. PMID 10704475.
- ^ a b c m Prescott L (1993). Microbiology. Wm. C. Brown Publishers. ISBN 0-697-01372-3.
- ^ a b Bunting J (1997). "The Virus that Cures". BBC Horizon. BBC Worldwide Ltd. OCLC 224991186. – Documentary about the history of phage medicine in Russia and the West
- ^ Borrell B, Fishchetti V (August 2012). "Science talk: Phage factor". Scientific American. pp. 80–83. JSTOR 26016042.
- ^ Kortright KE, Chan BK, Koff JL, Turner PE (February 2019). "Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria". Cell Host & Microbe. 25 (2): 219–232. doi:10.1016/j.chom.2019.01.014. PMID 30763536. S2CID 73439131.
- ^ Gordillo Altamirano FL, Barr JJ (April 2019). "Phage Therapy in the Postantibiotic Era". Clinical Microbiology Reviews. 32 (2). doi:10.1128/CMR.00066-18. PMC 6431132. PMID 30651225.
- ^ González-Mora A, Hernández-Pérez J, Iqbal HM, Rito-Palomares M, Benavides J (September 2020). "Bacteriophage-Based Vaccines: A Potent Approach for Antigen Delivery". Vaccines. 8 (3): 504. doi:10.3390/vaccines8030504. PMC 7565293. PMID 32899720.
- ^ Keen EC (2012). "Phage therapy: concept to cure". Frontiers in Microbiology. 3: 238. doi:10.3389/fmicb.2012.00238. PMC 3400130. PMID 22833738.
- ^ a b c Popescu M, Van Belleghem JD, Khosravi A, Bollyky PL (September 2021). "Bacteriophages and the Immune System". Annual Review of Virology. 8 (1): 415–435. doi:10.1146/annurev-virology-091919-074551. PMID 34014761.
- ^ Stone E, Campbell K, Grant I, McAuliffe O (June 2019). "Understanding and Exploiting Phage-Host Interactions". Viruses. 11 (6): 567. doi:10.3390/v11060567. PMC 6630733. PMID 31216787.
- ^ Laanto E, Mäntynen S, De Colibus L, Marjakangas J, Gillum A, Stuart DI, et al. (August 2017). "Virus found in a boreal lake links ssDNA and dsDNA viruses". Proceedings of the National Academy of Sciences of the United States of America. 114 (31): 8378–8383. Bibcode:2017PNAS..114.8378L. doi:10.1073/pnas.1703834114. PMC 5547622. PMID 28716906.
- ^ Krishnamurthy SR, Wang D (March 2018). "Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses". Virology. 516: 108–114. doi:10.1016/j.virol.2018.01.006. PMID 29346073.
- ^ Callanan J, Stockdale SR, Adriaenssens EM, Kuhn JH (January 2021). "Rename one class (Leviviricetes - formerly Allassoviricetes), rename one order (Norzivirales - formerly Levivirales), create one new order (Timlovirales), and expand the class to a total of six families, 420 genera and 883 species". ResearchGate. doi:10.13140/RG.2.2.25363.40481.
- ^ Hankin EH (1896). "L'action bactericide des eaux de la Jumna et du Gange sur le vibrion du cholera". Annales de l'Institut Pasteur (in French). 10: 511–23.
- ^ Twort FW (1915). "An Investigation on the Nature of Ultra-Microscopic Viruses". The Lancet. 186 (4814): 1241–43. doi:10.1016/S0140-6736(01)20383-3.
- ^ d'Hérelles F (1917). "Sur un microbe invisible antagoniste des bacilles dysentériques" (PDF). Comptes Rendus de l'Académie des Sciences de Paris. 165: 373–5. Archived (PDF) from the original on 11 May 2011. Retrieved 5 September 2010.
- ^ d'Hérelles F (1949). "The bacteriophage" (PDF). Science News. 14: 44–59. Retrieved 5 September 2010.
- ^ Keen EC (December 2012). "Felix d'Herelle and our microbial future". Future Microbiology. 7 (12): 1337–1339. doi:10.2217/fmb.12.115. PMID 23231482.
- ^ Aswani VH, Shukla SK (June 2021). "An Early History of Phage Therapy in the United States: Is it Time to Reconsider?". Clinical Medicine & Research. 19 (2): 82–89. doi:10.3121/cmr.2021.1605. PMC 8231696. PMID 34172535.
- ^ "The Nobel Prize in Physiology or Medicine 1969". Nobel Foundation. Retrieved 28 July 2007.
- ^ Kuchment A (2012), The Forgotten Cure: The past and future of phage therapy, Springer, p. 11, ISBN 978-1-4614-0250-3
- ^ Myelnikov D (October 2018). "An Alternative Cure: The Adoption and Survival of Bacteriophage Therapy in the USSR, 1922-1955". Journal of the History of Medicine and Allied Sciences. 73 (4): 385–411. doi:10.1093/jhmas/jry024. PMC 6203130. PMID 30312428.
- ^ Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, Kuhl S, et al. (January 2010). "Phage therapy in clinical practice: treatment of human infections". Current Pharmaceutical Biotechnology. 11 (1): 69–86. doi:10.2174/138920110790725401. PMID 20214609. S2CID 31626252.
- ^ Golovin S (2017). "Бактериофаги: убийцы в роли спасителей" [Bacteriophages: killers as saviors]. Наука и жизнь [Nauka I Zhizn (Science and life)] (in Russian) (6): 26–33.
- ^ a b Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A (June 2009). "Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial". Journal of Wound Care. 18 (6): 237–8, 240–3. doi:10.12968/jowc.2009.18.6.42801. PMID 19661847.
- ^ a b c Wright A, Hawkins CH, Anggård EE, Harper DR (August 2009). "A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy". Clinical Otolaryngology. 34 (4): 349–357. doi:10.1111/j.1749-4486.2009.01973.x. PMID 19673983.
- ^ Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, Sunkari V, et al. (March 2019). "Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection". Science. 363 (6434): eaat9691. doi:10.1126/science.aat9691. PMC 6656896. PMID 30923196.
- ^ Tawil N, Sacher E, Mandeville R, Meunier M (May 2012). "Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages" (PDF). Biosensors & Bioelectronics. 37 (1): 24–29. doi:10.1016/j.bios.2012.04.048. PMID 22609555. Archived (PDF) from the original on 2 February 2023.
- ^ Cha K, Oh HK, Jang JY, Jo Y, Kim WK, Ha GU, et al. (10 April 2018). "Characterization of Two Novel Bacteriophages Infecting Multidrug-Resistant (MDR) Acinetobacter baumannii and Evaluation of Their Therapeutic Efficacy in Vivo". Frontiers in Microbiology. 9: 696. doi:10.3389/fmicb.2018.00696. PMC 5932359. PMID 29755420.
- ^ a b Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, et al. (October 2017). "Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection". Antimicrobial Agents and Chemotherapy. 61 (10). doi:10.1128/AAC.00954-17. PMC 5610518. PMID 28807909.
- ^ a b O'Sullivan L, Bolton D, McAuliffe O, Coffey A (March 2019). "Bacteriophages in Food Applications: From Foe to Friend". Annual Review of Food Science and Technology. 10 (1). Annual Reviews: 151–172. doi:10.1146/annurev-food-032818-121747. PMID 30633564. S2CID 58620015.
- ^ U.S. FDA/CFSAN: Agency Response Letter, GRAS Notice No. 000198
- ^ (U.S. FDA/CFSAN: Agency Response Letter, GRAS Notice No. 000218)
- ^ "FSIS Directive 7120: Safe and Suitable Ingredients Used in the Production of Meat, Poultry, and Egg Products" (PDF). Food Safety and Inspection Service. Washington, DC: United States Department of Agriculture. Archived from the original (PDF) on 18 October 2011.
- ^ Khan FM, Chen JH, Zhang R, Liu B (2023). "A comprehensive review of the applications of bacteriophage-derived endolysins for foodborne bacterial pathogens and food safety: recent advances, challenges, and future perspective". Frontiers in Microbiology. 14: 1259210. doi:10.3389/fmicb.2023.1259210. PMC 10588457. PMID 37869651.
- ^ Chacón L, Barrantes K, Santamaría-Ulloa C, Solano MReyes L, Taylor LValiente C, Symonds EM, Achí R. 2020. A Somatic Coliphage Threshold Approach To Improve the Management of Activated Sludge Wastewater Treatment Plant Effluents in Resource-Limited Regions. Appl Environ Microbiol 86:e00616-20. https://doi.org/10.1128/AEM.00616-20/
- ^ "FDA 510(k) Premarket Notification". U.S. Food and Drug Administration.
- ^ Chacón L, Barrantes K, Santamaría-Ulloa C, Solano M, Reyes L, Taylor L, et al. (6 May 2011). "FDA clears first test to quickly diagnose and distinguish MRSA and MSSA". Applied and Environmental Microbiology. 86 (17). U.S. Food and Drug Administration. doi:10.1128/aem.00616-20. PMC 7440787. PMID 32591380. Archived from the original on 17 February 2015. Retrieved 17 February 2015.
{{cite journal}}: CS1 maint: bot: original URL status unknown (link) - ^ Vaisman D (25 May 2007). "Studying anthrax in a Soviet-era lab – with Western funding". The New York Times.
- ^ Dobozi-King M, Seo S, Kim JU, Young R, Cheng M, Kish LB (2005). "Rapid detection and identification of bacteria: SEnsing of Phage-Triggered Ion Cascade (SEPTIC)" (PDF). Journal of Biological Physics and Chemistry. 5: 3–7. doi:10.4024/1050501.jbpc.05.01. Archived from the original (PDF) on 26 September 2018. Retrieved 19 December 2016.
- ^ Smith GP, Petrenko VA (April 1997). "Phage Display". Chemical Reviews. 97 (2): 391–410. doi:10.1021/cr960065d. PMID 11848876.
- ^ Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, et al. (February 2004). "Antimicrobial drug discovery through bacteriophage genomics". Nature Biotechnology. 22 (2): 185–191. doi:10.1038/nbt932. PMID 14716317. S2CID 9905115.
- ^ "Technological background Phage-ligand technology". bioMérieux.
- ^ Keen EC (January 2014). "Tradeoffs in bacteriophage life histories". Bacteriophage. 4 (1): e28365. doi:10.4161/bact.28365. PMC 3942329. PMID 24616839.
- ^ Atamer Z, Samtlebe M, Neve H, J Heller K, Hinrichs J (16 July 2013). "Review: elimination of bacteriophages in whey and whey products". Frontiers in Microbiology. 4: 191. doi:10.3389/fmicb.2013.00191. PMC 3712493. PMID 23882262.
- ^ Coffey A, Ross RP (August 2002). "Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application". Antonie van Leeuwenhoek. 82 (1–4). Springer: 303–321. doi:10.1023/a:1020639717181. PMID 12369198. S2CID 7217985.
- ^ Fernández L, Escobedo S, Gutiérrez D, Portilla S, Martínez B, García P, et al. (November 2017). "Bacteriophages in the Dairy Environment: From Enemies to Allies". Antibiotics. 6 (4). [MDPI]: 27. doi:10.3390/antibiotics6040027. PMC 5745470. PMID 29117107.
- ^ Abedon ST (October 2019). "Look Who's Talking: T-Even Phage Lysis Inhibition, the Granddaddy of Virus-Virus Intercellular Communication Research". Viruses. 11 (10): 951. doi:10.3390/v11100951. PMC 6832632. PMID 31623057.
- ^ Mason KA, Losos JB, Singer SR, Raven PH, Johnson GB (2011). Biology. New York: McGraw-Hill. p. 533. ISBN 978-0-07-893649-4.
- ^ Mokrousov I (January 2009). "Corynebacterium diphtheriae: genome diversity, population structure and genotyping perspectives". Infection, Genetics and Evolution. 9 (1): 1–15. Bibcode:2009InfGE...9....1M. doi:10.1016/j.meegid.2008.09.011. PMID 19007916.
- ^ Charles RC, Ryan ET (October 2011). "Cholera in the 21st century". Current Opinion in Infectious Diseases. 24 (5): 472–477. doi:10.1097/QCO.0b013e32834a88af. PMID 21799407. S2CID 6907842.
- ^ Keen EC (December 2012). "Paradigms of pathogenesis: targeting the mobile genetic elements of disease". Frontiers in Cellular and Infection Microbiology. 2: 161. doi:10.3389/fcimb.2012.00161. PMC 3522046. PMID 23248780.
- ^ Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B (2015). "Bacteriophages and phage-derived proteins--application approaches". Current Medicinal Chemistry. 22 (14): 1757–1773. doi:10.2174/0929867322666150209152851. PMC 4468916. PMID 25666799.
- ^ Gabashvili IS, Khan SA, Hayes SJ, Serwer P (October 1997). "Polymorphism of bacteriophage T7". Journal of Molecular Biology. 273 (3): 658–667. doi:10.1006/jmbi.1997.1353. PMID 9356254.
- ^ Maghsoodi A, Chatterjee A, Andricioaei I, Perkins NC (December 2019). "How the phage T4 injection machinery works including energetics, forces, and dynamic pathway". Proceedings of the National Academy of Sciences of the United States of America. 116 (50): 25097–25105. Bibcode:2019PNAS..11625097M. doi:10.1073/pnas.1909298116. PMC 6911207. PMID 31767752.
- ^ Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, et al. (April 1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene". Nature. 260 (5551): 500–507. Bibcode:1976Natur.260..500F. doi:10.1038/260500a0. PMID 1264203. S2CID 4289674.
- ^ Mizuno CM, Guyomar C, Roux S, Lavigne R, Rodriguez-Valera F, Sullivan MB, et al. (February 2019). "Numerous cultivated and uncultivated viruses encode ribosomal proteins". Nature Communications. 10 (1): 752. Bibcode:2019NatCo..10..752M. doi:10.1038/s41467-019-08672-6. PMC 6375957. PMID 30765709.
- ^ Snustad DP (August 1968). "Dominance interactions in Escherichia coli cells mixedly infected with bacteriophage T4D wild-type and amber mutants and their possible implications as to type of gene-product function: catalytic vs. stoichiometric". Virology. 35 (4): 550–563. doi:10.1016/0042-6822(68)90285-7. PMID 4878023.
- ^ Floor E (February 1970). "Interaction of morphogenetic genes of bacteriophage T4". Journal of Molecular Biology. 47 (3): 293–306. doi:10.1016/0022-2836(70)90303-7. PMID 4907266.
- ^ Petrov AS, Harvey SC (July 2008). "Packaging double-helical DNA into viral capsids: structures, forces, and energetics". Biophysical Journal. 95 (2): 497–502. Bibcode:2008BpJ....95..497P. doi:10.1529/biophysj.108.131797. PMC 2440449. PMID 18487310.
- ^ Edgar RS Conditional lethals: in Phage and the Origins of Molecular Biology (2007) Edited by John Cairns, Gunther S. Stent, and James D. Watson, Cold Spring Harbor Laboratory of Quantitative Biology, Cold Spring Harbor, Long Island, New York ISBN 978-0879698003
- ^ Edgar B (October 2004). "The genome of bacteriophage T4: an archeological dig". Genetics. 168 (2): 575–82. doi:10.1093/genetics/168.2.575. PMC 1448817. PMID 15514035.
- ^ a b Epstein RH, Bolle A, Steinberg CM, Kellenberger E, Boy de la Tour E, Chevalley R, et al. (1963). "Physiological Studies of Conditional Lethal Mutants of Bacteriophage T4D". Cold Spring Harbor Symposia on Quantitative Biology. 28: 375–394. doi:10.1101/SQB.1963.028.01.053. ISSN 0091-7451.
- ^ Edgar RS, Lielausis I (April 1964). "Temperature-sensitive mutants of bacteriophage T4D: Their isolation and Characterization". Genetics. 49 (4): 649–62. doi:10.1093/genetics/49.4.649. PMC 1210603. PMID 14156925.
- ^ Henrot C, Petit MA (November 2022). "Signals triggering prophage induction in the gut microbiota". Molecular Microbiology. 118 (5): 494–502. doi:10.1111/mmi.14983. PMC 9827884. PMID 36164818. S2CID 252542284.
- ^ Callaway E (2017). "Do you speak virus? Phages caught sending chemical messages". Nature. doi:10.1038/nature.2017.21313.
- ^ Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, et al. (January 2017). "Communication between viruses guides lysis-lysogeny decisions". Nature. 541 (7638): 488–493. Bibcode:2017Natur.541..488E. doi:10.1038/nature21049. PMC 5378303. PMID 28099413.
- ^ Black LW, Thomas JA (2012). "Condensed Genome Structure". Viral Molecular Machines. Advances in Experimental Medicine and Biology. Vol. 726. Springer. pp. 469–87. doi:10.1007/978-1-4614-0980-9_21. ISBN 978-1-4614-0979-3. PMC 3559133. PMID 22297527.
- ^ Al-Shayeb B, Sachdeva R, Chen LX, Ward F, Munk P, Devoto A, et al. (February 2020). "Clades of huge phages from across Earth's ecosystems". Nature. 578 (7795): 425–431. Bibcode:2020Natur.578..425A. doi:10.1038/s41586-020-2007-4. PMC 7162821. PMID 32051592.
- ^ Häuser R, Blasche S, Dokland T, Haggård-Ljungquist E, von Brunn A, Salas M, et al. (2012). "Bacteriophage protein-protein interactions". Advances in Virus Research. 83: 219–298. doi:10.1016/B978-0-12-394438-2.00006-2. ISBN 978-0-12-394438-2. PMC 3461333. PMID 22748812.
- ^ Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF (March 2008). "Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination". Journal of Bacteriology. 190 (6): 2172–2182. doi:10.1128/JB.01657-07. PMC 2258872. PMID 18178732.
- ^ Krupovic M, Prangishvili D, Hendrix RW, Bamford DH (December 2011). "Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere". Microbiology and Molecular Biology Reviews. 75 (4): 610–635. doi:10.1128/MMBR.00011-11. PMC 3232739. PMID 22126996.
- ^ Takahashi I, Marmur J (February 1963). "Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis". Nature. 197 (4869): 794–795. Bibcode:1963Natur.197..794T. doi:10.1038/197794a0. PMID 13980287. S2CID 4166988.
- ^ Kirnos MD, Khudyakov IY, Alexandrushkina NI, Vanyushin BF (November 1977). "2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA". Nature. 270 (5635): 369–370. Bibcode:1977Natur.270..369K. doi:10.1038/270369a0. PMID 413053. S2CID 4177449.
- ^ a b Klipp E (2009). Systems biology: a textbook. Weinheim: Wiley-VCH. ISBN 978-3-527-31874-2. OCLC 288986435.
- ^ Zhao X, Chen C, Shen W, Huang G, Le S, Lu S, et al. (January 2016). "Global Transcriptomic Analysis of Interactions between Pseudomonas aeruginosa and Bacteriophage PaP3". Scientific Reports. 6: 19237. Bibcode:2016NatSR...619237Z. doi:10.1038/srep19237. PMC 4707531. PMID 26750429.
- ^ Blasche S, Wuchty S, Rajagopala SV, Uetz P (December 2013). "The protein interaction network of bacteriophage lambda with its host, Escherichia coli". Journal of Virology. 87 (23): 12745–12755. doi:10.1128/JVI.02495-13. PMC 3838138. PMID 24049175.
- ^ Bobonis J, Mitosch K, Mateus A, Karcher N, Kritikos G, Selkrig J, et al. (September 2022). "Bacterial retrons encode phage-defending tripartite toxin-antitoxin systems". Nature. 609 (7925): 144–150. Bibcode:2022Natur.609..144B. doi:10.1038/s41586-022-05091-4. PMID 35850148. S2CID 250643138.
- ^ Ka D, Oh H, Park E, Kim JH, Bae E (June 2020). "Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+ degradation". Nature Communications. 11 (1): 2816. Bibcode:2020NatCo..11.2816K. doi:10.1038/s41467-020-16703-w. PMC 7272460. PMID 32499527.
- ^ a b Cieślik M, Bagińska N, Jończyk-Matysiak E, Węgrzyn A, Węgrzyn G, Górski A (May 2021). "Temperate Bacteriophages-The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions". Viruses. 13 (6): 1013. doi:10.3390/v13061013. PMC 8228536. PMID 34071422.
- ^ a b Wendling CC, Refardt D, Hall AR (February 2021). "Fitness benefits to bacteria of carrying prophages and prophage-encoded antibiotic-resistance genes peak in different environments". Evolution; International Journal of Organic Evolution. 75 (2): 515–528. doi:10.1111/evo.14153. PMC 7986917. PMID 33347602.
- ^ Kirsch JM, Brzozowski RS, Faith D, Round JL, Secor PR, Duerkop BA (September 2021). "Bacteriophage-Bacteria Interactions in the Gut: From Invertebrates to Mammals". Annual Review of Virology. 8 (1): 95–113. doi:10.1146/annurev-virology-091919-101238. PMC 8484061. PMID 34255542.
- ^ Brenciani A, Bacciaglia A, Vignaroli C, Pugnaloni A, Varaldo PE, Giovanetti E (January 2010). "Phim46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes". Antimicrobial Agents and Chemotherapy. 54 (1): 221–229. doi:10.1128/AAC.00499-09. PMC 2798480. PMID 19858262.
- ^ Jahn MT, Arkhipova K, Markert SM, Stigloher C, Lachnit T, Pita L, et al. (October 2019). "A Phage Protein Aids Bacterial Symbionts in Eukaryote Immune Evasion". Cell Host & Microbe. 26 (4): 542–550.e5. doi:10.1016/j.chom.2019.08.019. PMID 31561965. S2CID 203580138.
- ^ Leigh BA (October 2019). "Cooperation among Conflict: Prophages Protect Bacteria from Phagocytosis". Cell Host & Microbe. 26 (4): 450–452. doi:10.1016/j.chom.2019.09.003. PMID 31600498. S2CID 204243652.
- ^ Ali Y, Koberg S, Heßner S, Sun X, Rabe B, Back A, et al. (2014). "Temperate Streptococcus thermophilus phages expressing superinfection exclusion proteins of the Ltp type". Frontiers in Microbiology. 5: 98. doi:10.3389/fmicb.2014.00098. PMC 3952083. PMID 24659988.
- ^ McGrath S, Fitzgerald GF, van Sinderen D (January 2002). "Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages". Molecular Microbiology. 43 (2): 509–520. doi:10.1046/j.1365-2958.2002.02763.x. PMID 11985726. S2CID 7084706.
- ^ Douwe M, McGrath J, Fitzgerald S, van Sinderen GF. Identification and Characterization of Lactococcal-Prophage-Carried Superinfection Exclusion Genes▿ †. American Society for Microbiology (ASM). OCLC 679550931.
- ^ Brüssow H, Canchaya C, Hardt WD (September 2004). "Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion". Microbiology and Molecular Biology Reviews. 68 (3): 560–602, table of contents. doi:10.1128/MMBR.68.3.560-602.2004. PMC 515249. PMID 15353570.
- ^ Edlin G, Lin L, Kudrna R (June 1975). "Lambda lysogens of E. coli reproduce more rapidly than non-lysogens". Nature. 255 (5511): 735–737. Bibcode:1975Natur.255..735E. doi:10.1038/255735a0. PMID 1094307. S2CID 4156346.
- ^ Sekulovic O, Fortier LC (February 2015). Schaffner DW (ed.). "Global transcriptional response of Clostridium difficile carrying the CD38 prophage". Applied and Environmental Microbiology. 81 (4): 1364–1374. Bibcode:2015ApEnM..81.1364S. doi:10.1128/AEM.03656-14. PMC 4309704. PMID 25501487.
- ^ Rossmann FS, Racek T, Wobser D, Puchalka J, Rabener EM, Reiger M, et al. (February 2015). "Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing". PLOS Pathogens. 11 (2): e1004653. doi:10.1371/journal.ppat.1004653. PMC 4338201. PMID 25706310.
- ^ Obeng N, Pratama AA, Elsas JD (June 2016). "The Significance of Mutualistic Phages for Bacterial Ecology and Evolution" (PDF). Trends in Microbiology. 24 (6): 440–449. doi:10.1016/j.tim.2015.12.009. PMID 26826796. S2CID 3565635.
- ^ Li G, Cortez MH, Dushoff J, Weitz JS (July 2020). "When to be temperate: on the fitness benefits of lysis vs. lysogeny". Virus Evolution. 6 (2): veaa042. bioRxiv 10.1101/709758. doi:10.1093/ve/veaa042. PMC 9532926. PMID 36204422.
- ^ Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, et al. (October 2002). "Genomic analysis of uncultured marine viral communities". Proceedings of the National Academy of Sciences of the United States of America. 99 (22): 14250–14255. Bibcode:2002PNAS...9914250B. doi:10.1073/pnas.202488399. PMC 137870. PMID 12384570.
- ^ Martin C (1988). "The Application of Bacteriophage Tracer Techniques in South West Water". Water and Environment Journal. 2 (6): 638–642. Bibcode:1988WaEnJ...2..638M. doi:10.1111/j.1747-6593.1988.tb01352.x.
- ^ Bergh O, Børsheim KY, Bratbak G, Heldal M (August 1989). "High abundance of viruses found in aquatic environments". Nature. 340 (6233): 467–468. Bibcode:1989Natur.340..467B. doi:10.1038/340467a0. PMID 2755508. S2CID 4271861.
- ^ Keen EC, Bliskovsky VV, Malagon F, Baker JD, Prince JS, Klaus JS, et al. (January 2017). "Novel "Superspreader" Bacteriophages Promote Horizontal Gene Transfer by Transformation". mBio. 8 (1): e02115–16. doi:10.1128/mBio.02115-16. PMC 5241400. PMID 28096488.
- ^ Lekunberri I, Subirats J, Borrego CM, Balcázar JL (January 2017). "Exploring the contribution of bacteriophages to antibiotic resistance". Environmental Pollution. 220 (Pt B): 981–984. Bibcode:2017EPoll.220..981L. doi:10.1016/j.envpol.2016.11.059. hdl:10256/14115. PMID 27890586.
- ^ Beavogui A, Lacroix A, Wiart N, Poulain J, Delmont TO, Paoli L, et al. (March 2024). "The defensome of complex bacterial communities". Nature Communications. 15 (1): 2146. Bibcode:2024NatCo..15.2146B. doi:10.1038/s41467-024-46489-0. PMC 10924106. PMID 38459056.
- ^ a b Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ (September 2016). "Healthy human gut phageome". Proceedings of the National Academy of Sciences of the United States of America. 113 (37): 10400–10405. Bibcode:2016PNAS..11310400M. doi:10.1073/pnas.1601060113. PMC 5027468. PMID 27573828.
- ^ Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. (October 2011). "The human gut virome: inter-individual variation and dynamic response to diet". Genome Research. 21 (10): 1616–1625. doi:10.1101/gr.122705.111. PMC 3202279. PMID 21880779.
- ^ a b c Kirsch JM, Brzozowski RS, Faith D, Round JL, Secor PR, Duerkop BA (September 2021). "Bacteriophage-Bacteria Interactions in the Gut: From Invertebrates to Mammals". Annual Review of Virology. 8 (1): 95–113. doi:10.1146/annurev-virology-091919-101238. PMC 8484061. PMID 34255542.
- ^ Strauss JH, Sinsheimer RL (July 1963). "Purification and properties of bacteriophage MS2 and of its ribonucleic acid". Journal of Molecular Biology. 7 (1): 43–54. doi:10.1016/S0022-2836(63)80017-0. PMID 13978804.
- ^ Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W (March 2003). "Bacteriophage T4 genome". Microbiology and Molecular Biology Reviews. 67 (1): 86–156, table of contents. doi:10.1128/MMBR.67.1.86-156.2003. PMC 150520. PMID 12626685.
- ^ Ackermann HW, Krisch HM (6 April 2014). "A catalogue of T4-type bacteriophages". Archives of Virology. 142 (12): 2329–2345. doi:10.1007/s007050050246. PMID 9672598. S2CID 39369249.
- ^ Wang RH, Yang S, Liu Z, Zhang Y, Wang X, Xu Z, et al. (January 2024). "PhageScope: a well-annotated bacteriophage database with automatic analyses and visualizations". Nucleic Acids Research. 52 (D1): D756–D761. doi:10.1093/nar/gkad979. PMC 10767790. PMID 37904614.
Bibliography
edit- Hauser AR, Mecsas J, Moir DT (July 2016). "Beyond Antibiotics: New Therapeutic Approaches for Bacterial Infections". Clinical Infectious Diseases. 63 (1): 89–95. doi:10.1093/cid/ciw200. PMC 4901866. PMID 27025826.
- Strathdee S, Patterson T (2019). The Perfect Predator. Hachette Books. ISBN 978-0-316-41808-9.
- Häusler T (2006). Viruses vs. superbugs : a solution to the antibiotics crisis?. London: Macmillan. ISBN 978-1-4039-8764-8.
External links
edit- Abedon ST. "The Bacteriophage Ecology Group". The Ohio State University. Archived from the original on 3 June 2013.
- Tourterel C, Blouin Y. "Bacteriophages illustrations and genomics". Orsay phage web site. Archived from the original on 29 October 2013. Retrieved 24 October 2013.
- "QuipStories: Bacteriophages get a foothold on their prey" (PDF). PDBe.
- Flatow I (April 2008). "Using 'Phage' Viruses to Help Fight Infection". Science Friday podcast. NPR. Archived from the original on 17 April 2008.
- "Animation of a scientifically correct T4 bacteriophage targeting E. coli bacteria". YouTube. 21 May 2019.
- "T4 Bacteriophage targeting E. coli bacteria". Animation by Hybrid Animation Medical. 21 December 2009.
- Bacteriophages: What are they. Presentation by Professor Graham Hatfull, University of Pittsburgh on YouTube