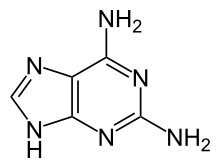
2,6-diaminopurine (2,6-DAP, also known as 2-aminoadenine) is a compound once used in the treatment of leukemia.[1] As the Z base, it is found instead of adenine (A) in the genetic material of some bacteriophage viruses.[2]
| |
| Names | |
|---|---|
| IUPAC name
7H-purine-2,6-diamine
| |
| Other names
2-aminoadenine; 2,6-DAP
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.016.006 |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C5H6N6 | |
| Molar mass | 150.145 g·mol−1 |
| Appearance | White to yellow crystalline powder |
| Density | 1.743 g/cm3 |
| Melting point | 117 to 122 °C (243 to 252 °F; 390 to 395 K) |
| 2.38 g/L at 20 °C | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
In August 2011, a report, based on NASA studies with meteorites found on Earth, was published suggesting 2,6-diaminopurine and related organic molecules, including the DNA and RNA components adenine and guanine, may have been formed extraterrestrially in outer space.[3][4][5]
In viruses
editIn cyanophage S-2L (Siphoviridae), diaminopurine is used instead of adenine (host evasion).[6] Diaminopurine base (Z) pairs perfectly with thymine (T) as it is identical to adenine (A) but has an amine group at position 2 forming 3 intermolecular hydrogen bonds, eliminating the major difference between the two types of basepairs (weak:A-T and strong:C-G). This improved stability affects protein-binding interactions that rely on those differences.
Four papers published April 2021 further describes the use and production of the Z-base. It is now known that:[7]
- The S-2L phage avoids incorporating A bases in the genome by hydrolyzing dATP (DatZ enzyme);[8]
- The Z base is produced by a pathway involving DUF550 (MazZ) and PurZ in S-2L and Vibrio phage PhiVC8;[9]
- The PrimPol/AEP DNA polymerase responsible for handling the Z base occurs in the same gene cluster as the three aforementioned enzymes;[10]
- The Z base is quite widespread in both Siphoviridae and Podoviridae, based on the occurrence of the said gene cluster.[11]
In August 2021, it was shown that DatZ, MazZ and PurZ are sufficient to replace some occurrence of A by Z in the bacterial genome of E. coli; expression of this system is toxic to the cell. The structures of MazZ (subtype 2) and PurZ are also determined, showing a possible link between PurZ and archaeal versions of PurA.[12]
Biosynthesis
edit2-aminoadenine is produced in two steps. The enzyme MazZ (homologous to MazG, EC 3.6.1.8) first performs:[12]
- dGTP + H2O = dGMP + diphosphate
The enzyme PurZ (homologous to PurA, EC 6.3.4.4) then performs:[9]
- (d)ATP + dGMP + L-aspartate = (d)ADP + phosphate + 2-aminodeoxyadenylosuccinate (dSMP)
The resulting dSMP is processed by host enzymes analogously to adenylosuccinate to produce dZTP.
In cellular life
editThis article is missing information about results of the altered H-bond strength in DNA and RNA. (October 2021) |
2,6-DAP was used to treat leukemia since as early as 1951.[13] It is known to arrest progression of cell cycle in mouse leukemia cells by 1989.[14] Cancer cells are known to become resistant to DAP by losing their adenine phosphoribosyltransferase (APRT) function,[15] a process shared with E. coli.[16]
DAP derivatives are in vitro antivirals useful against pseudorabies virus, a economically important livestock disease.[17] This base, in its free form, is able to correct UGA nonsense mutations by encouraging translational readthrough, through the inhibition of FTSJ1.[18]
Bioengineering
editIn bioengineering, anti-miRNA oligonucleotides (specifically, the serinol nucleic acid [SNA] type) incorporating base Z instead of A show enhanced binding to RNA.[19]
DAP is used similarly to other nuclear acid analogues in the investigation of enzyme structures and mechanisms.[20]
References
edit- ^ "George H. Hitchings". nobelprize.org.
- ^ "Some viruses thwart bacterial defenses with a unique genetic alphabet". 5 May 2021.
- ^ Callahan, M.P.; Smith, K.E.; Cleaves, H.J.; Ruzica, J.; Stern, J.C.; Glavin, D.P.; House, C.H.; Dworkin, J.P. (11 August 2011). "Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases". Proceedings of the National Academy of Sciences. 108 (34). PNAS: 13995–13998. Bibcode:2011PNAS..10813995C. doi:10.1073/pnas.1106493108. PMC 3161613. PMID 21836052.
- ^ Steigerwald, John (8 August 2011). "NASA Researchers: DNA Building Blocks Can Be Made in Space". NASA. Retrieved 2011-08-10.
- ^ ScienceDaily Staff (9 August 2011). "DNA Building Blocks Can Be Made in Space, NASA Evidence Suggests". ScienceDaily. Retrieved 2011-08-09.
- ^ Kirnos, MD; Khudyakov, IY; Alexandrushkina, NI; Vanyushin, BF (November 1977). "2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA". Nature. 270 (5635): 369–70. Bibcode:1977Natur.270..369K. doi:10.1038/270369a0. PMID 413053.
- ^ Bowler, Jacinta (4 May 2021). "Some Viruses Have a Completely Different Genome to The Rest of Life on Earth". ScienceAlert.
- ^ Czernecki, Dariusz; Legrand, Pierre; Tekpinar, Mustafa; Rosario, Sandrine; Kaminski, Pierre-Alexandre; Delarue, Marc (2021-04-23). "How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA". Nature Communications. 12 (1): 2420. Bibcode:2021NatCo..12.2420C. doi:10.1038/s41467-021-22626-x. ISSN 2041-1723. PMC 8065100. PMID 33893297.
- ^ a b Sleiman, Dona; Garcia, Pierre Simon; Lagune, Marion; Loc’h, Jerome; Haouz, Ahmed; Taib, Najwa; Röthlisberger, Pascal; Gribaldo, Simonetta; Marlière, Philippe; Kaminski, Pierre Alexandre (30 April 2021). "A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes". Science. 372 (6541): 516–520. Bibcode:2021Sci...372..516S. doi:10.1126/science.abe6494. PMID 33926955. S2CID 233448787.
- ^ Pezo, Valerie; Jaziri, Faten; Bourguignon, Pierre-Yves; Louis, Dominique; Jacobs-Sera, Deborah; Rozenski, Jef; Pochet, Sylvie; Herdewijn, Piet; Hatfull, Graham F.; Kaminski, Pierre-Alexandre; Marliere, Philippe (30 April 2021). "Noncanonical DNA polymerization by aminoadenine-based siphoviruses". Science. 372 (6541): 520–524. Bibcode:2021Sci...372..520P. doi:10.1126/science.abe6542. PMID 33926956. S2CID 233448788.
- ^ Zhou, Yan; Su, Xuexia; Wei, Yifeng; Cheng, Yu; Guo, Yu; Khudyakov, Ivan; Liu, Fuli; He, Ping; Song, Zhangyue; Li, Zhi; Gao, Yan; Ang, Ee Lui; Zhao, Huimin; Zhang, Yan; Zhao, Suwen (30 April 2021). "A widespread pathway for substitution of adenine by diaminopurine in phage genomes". Science. 372 (6541): 512–516. Bibcode:2021Sci...372..512Z. doi:10.1126/science.abe4882. PMID 33926954. S2CID 233448821.
- ^ a b Czernecki, Dariusz; Bonhomme, Frédéric; Kaminski, Pierre-Alexandre; Delarue, Marc (2021-08-05). "Characterization of a triad of genes in cyanophage S-2L sufficient to replace adenine by 2-aminoadenine in bacterial DNA". Nature Communications. 12 (1): 4710. Bibcode:2021NatCo..12.4710C. bioRxiv 10.1101/2021.04.30.442174. doi:10.1038/s41467-021-25064-x. ISSN 2041-1723. PMC 8342488. PMID 34354070.
- ^ Burchenal, JH; Karnofsky, DA; Kingsley-Pillers, EM; Southam, CM; Myers, WP; Escher, GC; Craver, LF; Dargeon, HW; Rhoads, CP (May 1951). "The effects of the folic acid antagonists and 2,6-diaminopurine on neoplastic disease, with special reference to acute leukemia". Cancer. 4 (3): 549–69. doi:10.1002/1097-0142(195105)4:3<549::aid-cncr2820040308>3.0.co;2-j. PMID 14839611. S2CID 31262125.
- ^ Weckbecker, G; Cory, JG (1989). "Metabolic activation of 2,6-diaminopurine and 2,6-diaminopurine-2'-deoxyriboside to antitumor agents". Advances in Enzyme Regulation. 28: 125–44. doi:10.1016/0065-2571(89)90068-x. PMID 2624171.
- ^ Shao, C; Deng, L; Henegariu, O; Liang, L; Stambrook, PJ; Tischfield, JA (20 June 2000). "Chromosome instability contributes to loss of heterozygosity in mice lacking p53". Proceedings of the National Academy of Sciences of the United States of America. 97 (13): 7405–10. Bibcode:2000PNAS...97.7405S. doi:10.1073/pnas.97.13.7405. PMC 16558. PMID 10861008.
- ^ Kocharian, ShM; Chukanova, TI; Sukhodolets, VV (1977). "[Mutations of resistance to 2,6-diaminopurine and 6-methylpurine that affect adenine phosphoribosyltransferase in Escherichia coli K-12]". Genetika. 13 (10): 1821–30. PMID 348574.
- ^ Zouharova, D; Lipenska, I; Fojtikova, M; Kulich, P; Neca, J; Slany, M; Kovarcik, K; Turanek-Knotigova, P; Hubatka, F; Celechovska, H; Masek, J; Koudelka, S; Prochazka, L; Eyer, L; Plockova, J; Bartheldyova, E; Miller, AD; Ruzek, D; Raska, M; Janeba, Z; Turanek, J (29 February 2016). "Antiviral activities of 2,6-diaminopurine-based acyclic nucleoside phosphonates against herpesviruses: In vitro study results with pseudorabies virus (PrV, SuHV-1)". Veterinary Microbiology. 184: 84–93. doi:10.1016/j.vetmic.2016.01.010. PMID 26854349.
- ^ Trzaska, C; Amand, S; Bailly, C; Leroy, C; Marchand, V; Duvernois-Berthet, E; Saliou, JM; Benhabiles, H; Werkmeister, E; Chassat, T; Guilbert, R; Hannebique, D; Mouray, A; Copin, MC; Moreau, PA; Adriaenssens, E; Kulozik, A; Westhof, E; Tulasne, D; Motorin, Y; Rebuffat, S; Lejeune, F (20 March 2020). "2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations". Nature Communications. 11 (1): 1509. Bibcode:2020NatCo..11.1509T. doi:10.1038/s41467-020-15140-z. PMC 7083880. PMID 32198346.
- ^ Kamiya, Y; Donoshita, Y; Kamimoto, H; Murayama, K; Ariyoshi, J; Asanuma, H (5 October 2017). "Introduction of 2,6-Diaminopurines into Serinol Nucleic Acid Improves Anti-miRNA Performance". ChemBioChem. 18 (19): 1917–1922. doi:10.1002/cbic.201700272. PMID 28748559. S2CID 35619213.
- ^ Bailly, C (1 October 1998). "The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA". Nucleic Acids Research. 26 (19): 4309–4314. doi:10.1093/nar/26.19.4309. PMC 147870. PMID 9742229.